Figure 4.
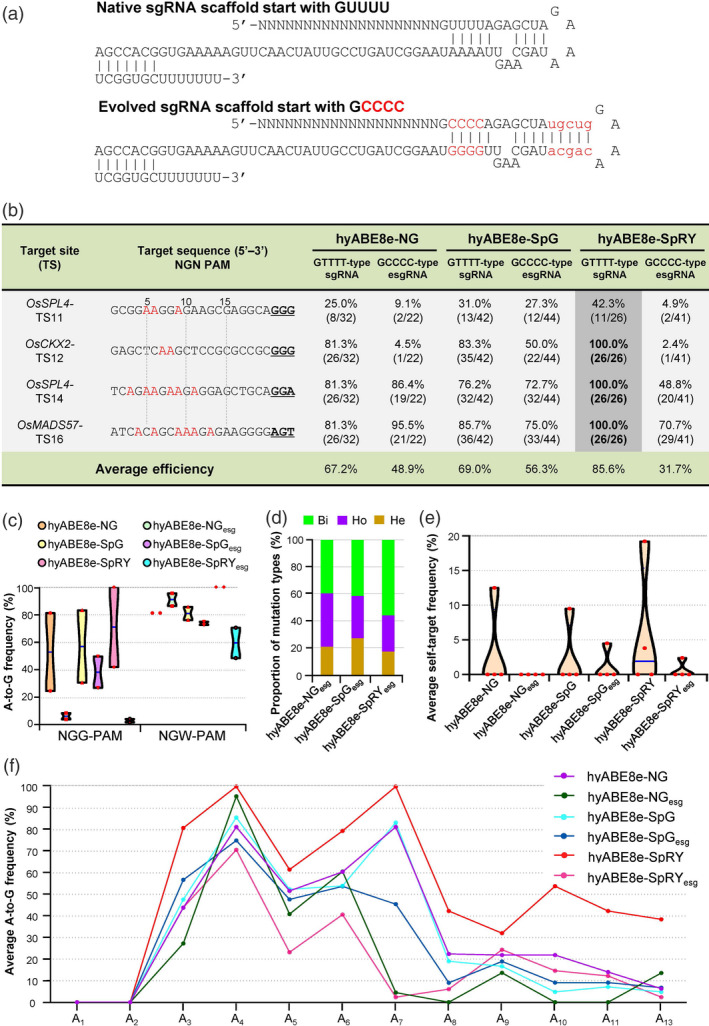
Evolved sgRNA esgRNA does not work well with PhieABEs. (a) Schematic diagram of the original sgRNA and evolved sgRNA (esgRNA). The esgRNA scaffold starting with ‘GCCCC’ and containing a longer stem domain was used in hyABE8e‐NG, hyABE8e‐SpG and hyABE8e‐SpRY. The replaced nucleotides are shown in red and uppercase font; the additional nucleotides are shown in red and lowercase font. (b) Comparison of sgRNA‐ and esgRNA‐guided base‐editing efficiency at TS11, TS12, TS14 and TS16 sites. GUUUU‐type sgRNA, native sgRNA; GCCCC‐type sgRNA, esgRNA (c) Editing efficiencies at NGG‐ and NGW‐PAM (where W is A or T) targets in the T0 plants edited by sgRNA‐ and esgRNA‐guided PhieABEs. PhieABEs with ‘esg’ (also in d–f) indicate results obtained with esgRNAs. (d) Proportion of mutation types induced by esgRNA‐guided PhieABEs. (e) Self‐targeted editing efficiencies in the sgRNA expression cassettes of esgRNA‐guided PhieABEs. Self‐targeted frequencies were much lower when esgRNAs were used, probably due to their lower editing activity with PhieABEs. (f) Editing activity windows and efficiencies with sgRNA‐ and esgRNA‐guided PhieABEs.
