Abstract
Background:
The heart grows in response to pathological and physiological stimuli. The former often precedes cardiomyocyte loss and heart failure; the latter paradoxically protects the heart and enhances cardiomyogenesis. The mechanisms underlying these differences remain incompletely understood. While long noncoding RNAs (lncRNAs) are important in cardiac development and disease, less is known about their roles in physiological hypertrophy or cardiomyogenesis.
Methods:
RNA sequencing was applied to hearts from mice after eight weeks voluntary exercise-induced physiological hypertrophy and cardiomyogenesis or transverse aortic constriction (TAC) for two or eight weeks to induce pathological hypertrophy or heart failure. The top lncRNA candidate was overexpressed in hearts with adeno-associated virus (AAV) vectors and inhibited with antisense locked nucleic acid (LNA)-GapmeRs to examine its function. Downstream effectors were identified through promoter analyses and binding assays. The functional roles of a novel downstream effector, dachsous cadherin-related 2 (DCHS2), were examined through transgenic overexpression in zebrafish and cardiac-specific deletion in Cas9-knockin mice.
Results:
We identified exercise-regulated cardiac lncRNAs, termed lncExACTs. lncExACT1 was evolutionarily conserved and decreased in exercised hearts but increased in human and experimental heart failure. Cardiac lncExACT1 overexpression caused pathological hypertrophy and heart failure, while lncExACT1 inhibition induced physiological hypertrophy and cardiomyogenesis, protecting against cardiac fibrosis and dysfunction. lncExACT1 functioned by regulating microRNA-222, calcineurin signaling, and Hippo/Yap1 signaling through DCHS2. Cardiomyocyte DCHS2 overexpression in zebrafish induced pathological hypertrophy and impaired cardiac regeneration, promoting scarring after injury. In contrast, murine DCHS2 deletion induced physiological hypertrophy and promoted cardiomyogenesis.
Conclusions:
These studies identify lncExACT1-DCHS2 as a novel pathway regulating cardiac hypertrophy and cardiomyogenesis. lncExACT1-DCHS2 acts as a master switch toggling the heart between physiological and pathological growth to determine functional outcomes, providing a potentially tractable therapeutic target for harnessing the beneficial effects of exercise.
Keywords: Long noncoding RNA, Cardiac hypertrophy, Exercise, Heart failure, Hippo/Yap1
Introduction
Heart failure (HF) is a growing cause of morbidity and mortality1. Prognosis remains poor for many patients with HF despite the best available treatments1. Physical activity is associated with a lower risk of HF2, although exercise training itself can induce cardiac hypertrophy. In animal models, exercise training paradoxically reduces the hypertrophy, dysfunction, and fibrosis seen in response to pathological stimuli3. Such observations suggest that although pathological and physiological hypertrophy appear similar, they may reflect fundamentally different underlying mechanisms that exist in dynamic tension. This model is supported by their distinct gene expression profiles, which diverge early and include sentinel differences in expression of markers such as ANP, BNP, PGC1α, C/EBPβ and myosin heavy chain (MHC) isoforms, often used to distinguish these states4, 5.
Others have suggested that the difference between physiological and pathological hypertrophy is simply one of degree6, recognizing that exercise is intermittent while disease states are generally constant. Some work has also suggested the transition from early, compensated hypertrophy to HF hinges on other factors, such as failure of angiogenesis needed to support growth of the myocardium7. Of course, these possibilities are not mutually exclusive.
Another central distinction is that pathological hypertrophy is associated with death of cardiomyocytes8, which are not replaced due to the limited regenerative capacity of the adult heart9. In contrast, exercise protects against cardiomyocyte death and induces cardiomyogenesis in the adult mammalian heart10. Identifying pathways capable of recapitulating these benefits of exercise with a feasible path to translation would have important clinical implications.
Using multi-isotope imaging mass spectrometry (MIMS), we demonstrated that eight weeks of voluntary wheel running increased cardiomyogenesis in adult mice almost 5-fold10. miR-222, an exercise-induced microRNA that mitigates adverse remodeling after ischemic injury11, was necessary for exercise-induced cardiomyogenesis10 and cardiac growth11. However, miR-222 overexpression was not sufficient to induce cardiomyogenesis or physiological hypertrophy in vivo11, suggesting the importance of other pathways.
Here we compared cardiac long noncoding RNAs (lncRNAs) altered in physiological cardiac hypertrophy versus pathological hypertrophy or HF. We identified a novel set of cardiac lncRNAs, termed lncExACTs (for long noncoding exercise-associated cardiac transcripts). The only lncExACTs significantly changed in both the physiological and pathological models changed in opposite directions. One of these, lncExACT1, was uniquely downregulated by exercise and upregulated in pathological hypertrophy and HF. Overexpression of lncExACT1 was sufficient to induce pathological hypertrophy and HF, while its inhibition recapitulated multiple exercise phenotypes, including physiological hypertrophy, increased markers of cardiomyogenesis, and protection against HF. Mechanistic studies revealed that lncExACT1 acts by binding miR-222 and modulating Hippo/Yap signaling through transcriptional regulation of its genomic neighbor, dachsous cadherin-related 2 (DCHS2). Inhibition of lncExACT1 in vivo by systemic delivery of antisense oligonucleotides effectively mediated these benefits, underscoring the potential translational implications of these observations.
Methods
The data, methods, and study materials used to conduct the research will be available from the corresponding author upon reasonable request.
Animals studies were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Massachusetts General Hospital (MGH) Institutional Animal Care and Use Committee. Human studies were approved by the Partners or Colorado Multicenter Institutional Review Boards. A detailed description of the methods and supporting data are available in the Online Data Supplement.
Statistical analysis
Data are presented as mean±SEM unless otherwise indicated and analyzed using GraphPad Prism 8 (GraphPad Software). Unpaired, two-tailed Student’s t-test or, when assessing multiple groups, one-way analysis of variance (ANOVA) with Tukey’s post hoc test, were used as indicated. In Figure 1E, pairwise Wilcoxon rank sum test with Bonferroni correction was used. In Figure 4K, repeated measures ANOVA was used. p<0.05 was considered significant.
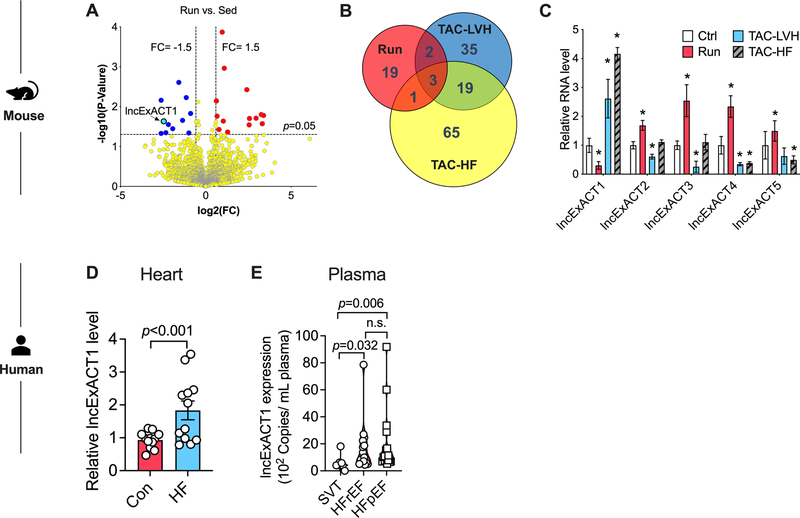
Figure 1. Identification of exercise-associated cardiac lncRNAs (lncExACTs).
A. Volcano plot showing lncRNA RNAseq results in hearts from exercised compared with sedentary mice, n=4 mice/group. B. Venn diagram of differentially regulated cardiac lncRNAs in exercised (Run) mice and animals with transverse aortic constriction (TAC)-induced left ventricular hypertrophy (TAC-LVH) or heart failure (TAC-HF), n=4 mice/group. C. QRT-PCR measurement of cardiac lncExACT1–5 expression in control (Ctrl), exercised (Run), TAC-LVH, or TAC-HF mice. *p<0.05 vs. Ctrl by one-way analysis of variance (ANOVA) with post hoc Tukey, n=4–5 mice/group. D. lncExACT1 expression measured by QRT-PCR is increased in hearts from heart failure (HF) patients compared with controls, n=12/group. p<0.001 by Student’s t test. E. Plasma lncExACT1 determined by ddPCR is increased in patients with HF with reduced ejection fraction (HFrEF, n=18) or HF with preserved ejection fraction (HFpEF, n=16) in comparison to supraventricular tachycardiac (SVT) patients without HF (n=8). p=0.006 and p=0.032 by pairwise Wilcoxon rank sum test with Bonferroni correction. Data shown as mean±SEM.
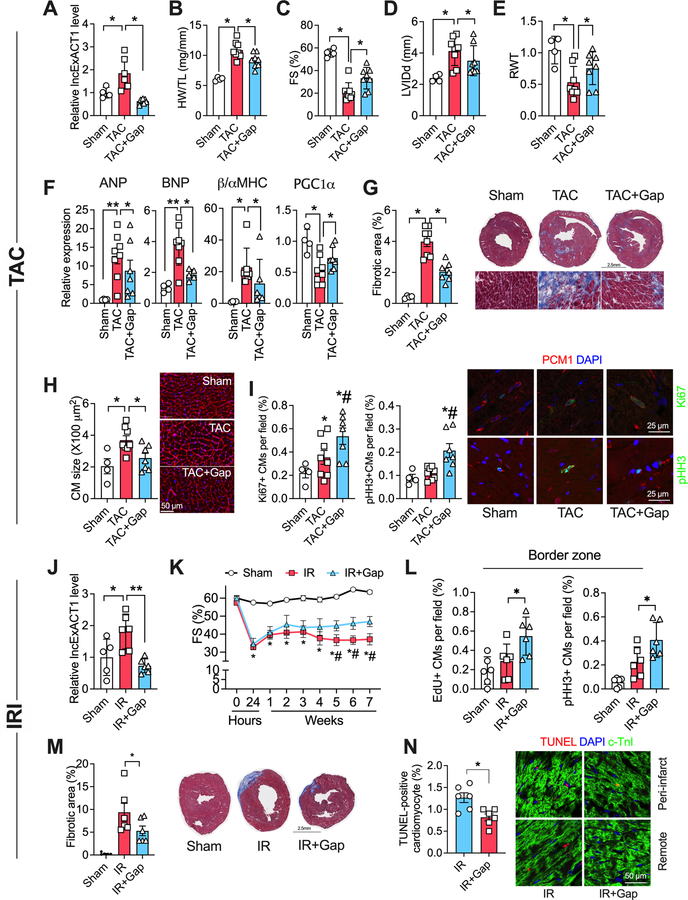
Figure 4. lncExACT1 inhibition protects against pathological hypertrophy, cardiac dysfunction, and fibrosis.
A. QRT-PCR measurement of lncExACT1 expression in hearts from mice subjected to sham operation (Sham), transverse aortic constriction with LNA-GapmeR-Control injection (TAC), or TAC with LNA-GapmeR-lncExACT1 (TAC+Gap). B Heart weight (HW) relative to tibial length (TL). C. Fractional shortening (FS). D. Left ventricular end diastolic internal dimension (LVIDd). E. Relative wall thickness (RWT). F. QRT-PCR measurement of hypertrophy markers in the heart. G. Quantification of fibrotic area from Masson trichrome-stained heart sections. H. Quantification of cardiomyocyte area from heart sections with wheat germ agglutinin (WGA) staining. I. Quantification of Ki67 and pHH3, and PCM1 double-positive cardiomyocytes in heart sections. J. QRT-PCR measurement of lncExACT1 expression in hearts from mice subjected to sham operation (Sham), myocardial ischemia reperfusion (IR) with LNA-GapmeR-control injection, or IR with LNA-GapmeR-lncExACT1 (IR+Gap). K. FS. L. Quantification of EdU and pHH3, and PCM1 double-positive cardiomyocytes at the infarct border zone in heart sections. M. Quantification of fibrotic area in Masson trichrome-stained heart sections. N. Quantification of TUNEL and cardiac troponin I (cTnI) double positive cardiomyocyte in heart sections. *p<0.05, **p<0.01; in I *p<0.05 vs. Sham, #p<0.05 vs. TAC by one-way analysis of variance (ANOVA) with post hoc Tukey. In K, *p<0.05 vs. Sham, #p<0.05 vs. IR+Gap by repeated repeated measures ANOVA. Data shown as mean±SEM.
Results
Identification and characterization of exercise-associated cardiac lncRNAs
RNAseq was performed on hearts from sedentary mice and mice subjected to voluntary running for 8 weeks, which induces physiological cardiac growth and cardiomyogenesis10. For comparison, RNAseq was also performed on hearts from mice subjected to transverse aortic constriction (TAC) for 2 or 8 weeks to induce pathological cardiac hypertrophy or HF, respectively. Principal Component Analysis (PCA) plots using all lncRNAs from the treatment groups (normalized to corresponding controls) showed lncRNAs clearly distinguish these states (Fig.I.A in Data Supplement). Twenty-five lncRNAs were differentially expressed (11 downregulated and 14 upregulated) in hearts from exercised compared with sedentary mice (Fig. 1A). We named these long noncoding Exercise-Associated Cardiac Transcripts (lncExACTs). Of these, six also changed in the pathological models; in each case in a direction opposite to that seen with exercise (Fig. 1B–C). One of these could not be reliably detected by quantitative reverse transcription polymerase chain reaction (QRT-PCR) but the other five all validated by QRT-PCR in independent cohorts (Fig. 1C). Among them, lncExACT1 was chosen for detailed study as the only lncExACT altered in both the physiological and pathological models that was downregulated in exercised hearts, suggesting antisense inhibition could have therapeutic benefits.
The full-length lncExACT1 sequence (ENSMUSG00000074517) was confirmed by rapid amplification of cDNA ends. lncExACT1 is highly conserved across mammalian species. The mouse ortholog of lncExACT1 exhibits 67% nucleotide identity with its human counterpart (NONHSAG111911) (Fig.I.B in Data Supplement). Of note, DCHS2 is the closest protein-coding gene in both species (overlapping in mice and within 136 bp in humans). The Coding Potential Assessment Tool (http://lilab.research.bcm.edu/cpat/) failed to identify any major open reading frames (ORF) with translational potential longer than 100 amino acids, however two small ORFs (sORFs, sORF1 is 306 base pairs (bp) and sORF2 is 525bp) were identified in lncExACT1. To test whether these sORFs have peptide-coding potential, full-length lncExACT1 or the sORFs were cloned upstream of the FLAG coding sequence in p3xFLAG-CMV and transfected into HEK293T cells. No FLAG-tagged peptides were detected (Fig.I.C in Data Supplement), suggesting these sORFs are not translated at a significant level. Moreover, constructs were generated in which a mutant GFP ORF (GFPmut, the start codon ATGGTT is mutated to ATTGTT) was fused to the 3’ end of the sORFs and the lncExACT1 sequence upstream of each sORF. Substantial expression of GFP fusion protein was observed in GFP wild-type but not in GFPmut, or GFPmut fused with sORF1 or sORF2. These results further argue against significant translation (Fig.I.D in Data Supplement), although very low translation levels cannot be completely excluded.
lncExACT1 is expressed by many tissues (Fig.I.E in Data Supplement), including the heart, in which expression is comparable in cardiomyocytes (35.59±3.60 copies/cell) and non-cardiomyocytes (28.23±4.60 copies/cell) (Fig.I.F in Data Supplement). While lncExACT1 is present in both cardiomyocyte nuclei and cytoplasm, it is ~2.5 times more abundant in the nucleus (Fig.I.G in Data Supplement). To see if lncExACT1 expression is affected by relevant stimuli, primary cardiomyocytes were exposed to IGF-1, which induces physiological cardiac growth through PI3-kinase12 and Akt13, 14. IGF-1 reduced lncExACT1 with a gene expression pattern consistent with physiological hypertrophy (Fig.I.H in Data Supplement). PI3-Kinase inhibition increased lncExACT1 and reversed the physiological expression pattern seen with IGF-1 treatment alone (Fig.I.H in Data Supplement), suggesting IGF-1 inhibits lncExACT1 through PI3-Kinase activation. In contrast, treatment with phenylephrine, which induces pathological hypertrophy through MAPKs15, increased lncExACT1 expression with a gene expression pattern consistent with pathological hypertrophy. These changes and lncExACT1 induction were attenuated by JNK inhibition (Fig.I.I-J in Data Supplement), while MEK and p38 inhibition did not block lncExACT1 induction.
In hearts explanted from patients with non-ischemic cardiomyopathy and reduced systolic function (ejection fraction (EF)=22.3±9.0%, n=12), lncExACT1 expression was increased ~1.8-fold in comparison to nonfailing (unused donor) hearts (EF=67.7±7.2%, n=12, p<10−11) from otherwise similar subjects (50% female and mean age 57 years for both, p>0.7) (Fig. 1D, p<0.001). In the entire population, cardiac lncExACT1 levels were weakly correlated with left ventricular EF (LVEF) but this association was driven by the lower levels in controls. In HF, lncExACT1 expression varied for unclear reasons and did not correlate with pre-explant LVEF (Fig.I.K–L in Data Supplement). In separate cohorts, we examined circulating lncExACT1 levels by Droplet Digital PCR (ddPCR) in plasma from patients with HF with reduced ejection fraction (HFrEF, n=16, EF=22.3±8.92%, p<10−14) and HF with preserved ejection fraction (HFpEF, n=18, EF=64.7±7.76%) in comparison to plasma from patients without HF and structurally normal hearts who presented with supraventricular tachycardia (SVT, n=8, EF=60.8±2.3%). Circulating lncExACT1 increased 2.9-fold in plasma from HFrEF patients (p=0.032 vs SVT) and 3.4-fold in HFpEF patients (p=0.006 vs SVT) (Fig. 1E). Together these data underscore the potential clinical relevance of lncExACT1.
lncExACT1 overexpression induces pathological cardiac hypertrophy
In primary cardiomyocytes, lentiviral expression of lncExACT1 increased cardiomyocyte size and induced a gene expression pattern characteristic of pathological hypertrophy including increased ANP, BNP, β/αMHC ratio, C/EBPβ, and decreased PGC1α (Fig.II.A–C in Data Supplement). Overexpression of lncExACT1 did not alter cardiomyocyte proliferation but enhanced proliferation of non-cardiomyocytes, predominantly made up of fibroblasts (Fig.II.D in Data Supplement). In primary cardiomyocytes, lentiviral expression of lncExACT1 decreased DSCR1, an endogenous inhibitor of calcineurin, a well-established driver of pathological hypertrophy16, while increasing protein expression of CnA (the catalytic subunit of calcineurin) and nuclear NFATc3 (downstream effector of calcineurin) (Fig.II.E in Data Supplement). Inhibition of calcineurin with FK506 partially attenuated lncExACT1-induced pathological gene expression (Fig.II.F in Data Supplement). These results suggest that lncExACT1 may induce pathological hypertrophy at least in part through activation of calcineurin signaling.
We next injected a cardiotropic adeno-associated virus (AAV9, 2*1012 GC/mouse) encoding lncExACT1 driven by a cardiac-specific promoter via the tail veins of wild-type mice. AAV9-lncExACT1 increased cardiac lncExACT1 gene expression ~7-fold at 16 weeks compared to control vector-injected mice (Fig. 2A), which is somewhat but not dramatically greater than the ~2–5-fold increase seen in human and experimental HF. AAV9-lncExACT1 increased expression both in the cytoplasm and in the nucleus (Fig.II.G in Data Supplement), where lncExACT1 is more abundant (Fig.II.H in Data Supplement). lncExACT1 expression was sufficient to increase heart weight relative to tibial length (HW/TL) and relative wall thickness (RWT) (Fig. 2B–C). Fractional shortening (FS) was reduced (Fig. 2D) consistent with impaired systolic function, and lung weight relative to tibial length (LW/TL) was modestly increased (Fig. 2E) consistent with HF. Of note, chamber size was reduced rather than increased as commonly seen in TAC-HF (Fig. 2F and Table I in Data Supplement). These changes were associated with a pathological gene expression pattern (Fig. 2G). lncExACT1 overexpression increased cardiomyocyte size but did not affect markers of cardiomyocyte proliferation (Fig. 2H–I and Fig.II.I–J in Data Supplement).
Figure 2. lncExACT1 overexpression induces pathological hypertrophy.
A. QRT-PCR measurement of lncExACT1 in mouse hearts 16 weeks after injection with AAV-lncExACT1 (lncExACT1) or AAV-GFP (Con). B Heart weight (HW) relative to tibial length (TL). C. Relative wall thickness (RWT). D. Fractional shortening (FS). E. Lung weight (LW) relative to tibial length (TL). F. Left ventricular end diastolic internal dimension (LVIDd). G. QRT-PCR measurement of hypertrophy markers in the heart. H. Quantification of cardiomyocyte area from wheat germ agglutinin (WGA)-stained heart sections. I. Quantification of EdU, PCM1 double-positive cardiomyocytes in heart sections. *p<0.05, **p<0.01 by Student’s t test. Data shown as mean±SEM.
Thus, lncExACT1 expression is sufficient to induce cardiac hypertrophy most consistent with pathological hypertrophy given the changes in gene expression and the development of cardiac dysfunction and HF. Moreover, there was no evidence of increased cardiomyogenesis, a hallmark of exercise-induced physiological hypertrophy10.
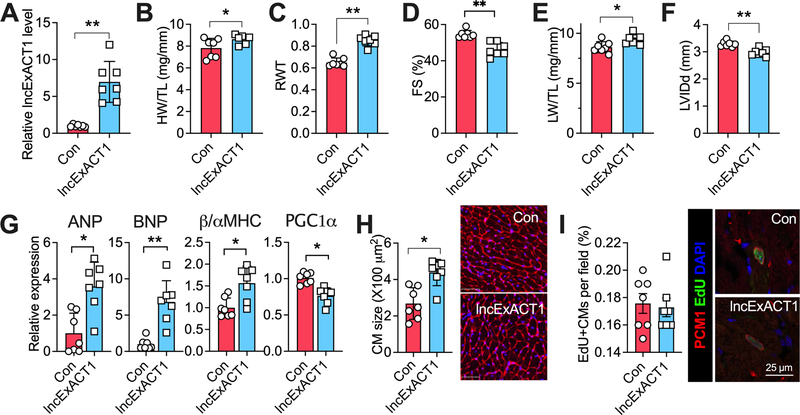
lncExACT1 inhibition induces physiological cardiac hypertrophy
Since lncExACT1 is reduced in the exercised heart, we next examined whether knockdown of lncExACT1 could mimic exercise-induced physiological cardiac growth. In primary cardiomyocytes, transfection with two locked nucleic acid (LNA) antisense oligonucleotides (GapmeR #1 and #2) specific to different regions of lncExACT1 each led to a ~50% reduction in lncExACT1 transcript levels (Fig.III.A in Data Supplement). Paradoxically, lncExACT1 knockdown increased cardiomyocyte size as did overexpression (Fig.III.B in Data Supplement). However, in contrast to overexpression, lncExACT1 knockdown induced a gene expression pattern consistent with physiological hypertrophy (Fig.III.C in Data Supplement). Inhibition of lncExACT1 increased the number of cardiomyocytes (Fig.III.B in Data Supplement) and increased markers of proliferation in cardiomyocytes while suppressing these markers in non-cardiomyocytes, predominantly fibroblasts (Fig.III.B,D–E in Data Supplement).
To examine effects of lncExACT1 inhibition in vivo, GapmeR #1 was injected via the tail vein into mice. Two weeks of lncExACT1 GapmeR treatment reduced cardiac lncExACT1 expression by ~50% (Fig. 3A), comparable to the ~70% reduction seen in exercised hearts (Fig. 1C). lncExACT1 GapmeR reduced lncExACT1 expression both in cardiomyocytes and fibroblasts, with higher efficiency in cardiomyocytes (Fig.III.F in Data Supplement). Inhibition of lncExACT1 increased HW/TL by ~23%, increased relative wall thickness by 24% (Fig. 3B–C), and improved cardiac function (Fig. 3D and Table II in Data Supplement), without affecting LW/TL or chamber dimensions (Fig. 3E–F), similar to the changes seen in exercised mice (data not shown). Inhibition of lncExACT1 induced a gene expression pattern most consistent with physiological hypertrophy (Fig. 3G). Inhibition of lncExACT1 increased cardiomyocyte size (Fig. 3H) comparably to lncExACT1 overexpression (Fig. 2H). However, lncExACT1 inhibition also increased markers of cardiomyocyte proliferation including increased 5-ethynyl-2´-deoxyuridine (EdU), Ki67, and phosphorylated histone H3 (pHH3) in cells also positive for pericentriolar material-1 (PCM1, a marker of cardiomyocyte nuclei17) (Fig. 3I–K). Taken together these data demonstrate that lncExACT1 inhibition is sufficient to induce physiological cardiac hypertrophy and markers of cardiomyogenesis, similar to those induced by 8 weeks of exercise.
Figure 3. lncExACT1 inhibition induces physiological hypertrophy.
A. QRT-PCR measurement of lncExACT1 in hearts from mice injected with LNA-GapmeR-Control (Con) or LNA-GapmeR-lncExACT1 (Gap) for 2 weeks. B. Heart weight (HW) relative to tibial length (TL). C. Relative wall thickness (RWT). D. Fractional shortening (FS). E. Lung weight (LW) relative to tibial length (TL). F. Left ventricular end diastolic internal dimension (LVIDd). G. QRT-PCR measurement of hypertrophy markers in the heart. H. Quantification of cardiomyocyte area from wheat germ agglutinin (WGA)-stained heart sections. I. Quantification of EdU, PCM1 double-positive cardiomyocytes in stained heart sections. J. Quantification of Ki67, PCM1 double-positive cardiomyocytes in stained heart sections. K. Quantification of pHH3, PCM1 double-positive cardiomyocytes in stained heart sections. *p<0.05, **p<0.01 by Student’s t test. Data shown as mean±SEM.
lncExACT1 inhibition protects the heart against adverse remodeling
Given that exercise protects the heart against pathological stress18, 19, we asked whether lncExACT1 inhibition protects against pressure-overload (TAC). Treatment with lncExACT1-specific GapmeR for 6 weeks completely blocked the TAC-induced increase in cardiac lncExACT1 (Fig. 4A). lncExACT1 inhibition also mitigated the TAC-induced increase in heart weight (Fig. 4B) and improved cardiac function, while reducing chamber dilatation and wall thinning (Fig. 4C–E and Table III in Data Supplement). lncExACT1 inhibition partially reversed pathological gene expression (Fig. 4F), while reducing cardiac fibrosis 2-fold and blocking the increase in cardiomyocyte size seen after TAC (Fig. 4G–H). Thus, although lncExACT1 inhibition induces physiological hypertrophy at baseline, it actually reduces pathological hypertrophy, suggesting a dynamic tension between physiological and pathological hypertrophy that is mediated by lncExACT1. Interestingly, Ki67 and pHH3, markers of proliferation, were modestly increased in cardiomyocytes after TAC, consistent with prior reports20, 21, and were further increased by lncExACT1 inhibition (Fig. 4I).
We next examined whether lncExACT1 inhibition mitigated adverse remodeling after ischemic injury. In mice subjected to myocardial ischemia reperfusion injury (IRI), injection of lncExACT1-specific GapmeR for 7 weeks starting after reperfusion reduced the IRI-induced increase in cardiac lncExACT1 in comparison to control GapmeR-injected mice (Fig. 4J–K). lncExACT1 inhibition increased markers of cardiomyocyte proliferation, EdU incorporation and pHH3 staining in the infarct border zone (Fig. 4L), and reduced cardiomyocyte apoptosis (Fig. 4N). lncExACT1 inhibition did not affect the initial injury as reflected in the reduced FS seen at 24 hours (Fig. 4K). However, the benefits of lncExACT1 inhibition became apparent after 4 weeks (Fig. 4K), suggesting improved initial cardiomyocyte survival may not be the dominant factor. Altogether, these results indicate that inhibition of lncExACT1 is sufficient to protect the heart against pathological hypertrophy, cardiac dysfunction, and adverse remodeling, likely through a combination of enhancing cardiomyocyte proliferation and survival culminating in dramatically reduced cardiac fibrosis.
lncExACT1 binds miR-222
lncRNAs can act as competitive endogenous RNAs (ceRNAs) binding miRNAs, typically in the cytoplasm22 to inhibit their actions. Bioinformatic analyses (http://starbase.sysu.edu.cn) identified five miRNAs as potentially binding lncExACT1 (Fig.IV.A in Data Supplement). We examined lncExACT1 binding in primary cardiomyocytes by QRT-PCR after pulldown of biotinylated probes specific for the sense or anti-sense strands of lncExACT1. Only miR-876–5p and miR-222–3p demonstrated detectable binding to lncExACT1. miR-876–5p, but not miR-222–3p, also bound lncExACT1 antisense though at a lower level (Fig.IV.B in Data Supplement). Sequences corresponding to the predicted wild-type or mutated miRNA binding sites of lncExACT1 were inserted 3’ of a luciferase reporter and co-transfected into cardiomyocytes with the related miRNA, miR-876–5p or miR-222–3p (Fig.IV.C in Data Supplement). The miR-222–3p mimic reduced luciferase activity of the construct with wild-type (wt) but not mutated (mut) putative miR-222 binding sites. In contrast, transfection with miR-876–5p did not affect luciferase activity with either the wild-type or mutated binding site (Fig.IV.D in Data Supplement). miR-222 inhibition reversed some of the physiological gene expression pattern otherwise seen with lncExACT1 knockdown and blocked the increase in cell size seen with lncExACT1 knockdown (Fig.IV.E–F in Data Supplement) without affecting lncExACT1 knockdown-induced cardiomyocyte proliferation (Fig.IV.G in Data Supplement). Taken together, these data suggest that miR-222, which we previously demonstrated is necessary for exercise-induced cardiac growth11, uniquely among the bioinformatic candidates binds lncExACT1 and contributes to the physiological cardiomyocyte hypertrophy but not the proliferation seen with lncExACT1 inhibition. Based on these observations as well as our prior work demonstrating even massive miR-222 overexpression is not sufficient to induce hypertrophy or cardiomyogenesis in the adult heart, we sought to identify other effectors contributing the phenotypes observed, particularly the changes in cardiomyocyte proliferation.
lncExACT1 regulates DCHS2 and Yap1
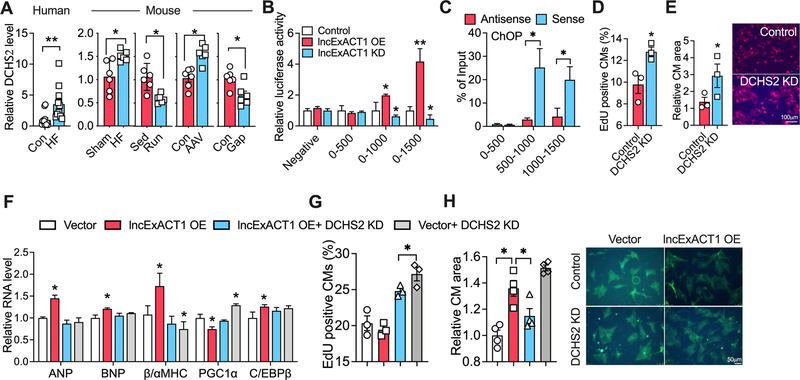
lncRNAs often regulate expression of nearby genes23, so we examined DCHS2, the protein-coding gene closest to lncExACT1 in both mice and humans. Paralleling lncExACT1, cardiac DCHS2 expression increased in patients and mice with HF but decreased in exercise (Fig. 5A). Moreover, cardiac DCHS2 decreased in mice after lncExACT1 knockdown and increased with AAV-lncExACT1 expression (Fig. 5A). Similar effects were seen in primary cardiomyocytes, while DCHS2 expression or knockdown did not affect lncExACT1 expression (Fig.V.A–B in Data Supplement). These data suggest DCHS2 expression may be regulated by lncExACT1. To determine if lncExACT1 directly regulates DCHS2 transcription, we performed analyses of the DCHS2 promoter. Sequences corresponding to 500, 1000, or 1500bp upstream of the DCHS2 transcriptional start site were inserted into a luciferase reporter and transfected into primary cardiomyocytes in combination with either lncExACT1 overexpression or inhibition. lncExACT1 expression increased while lncExACT1 knockdown decreased, the activity of promoter constructs that included 1000 or 1500 bp 5’ of DCHS2 but not the construct that only included 500 bp (Fig. 5B). Consistent with this, chromatin oligo-affinity precipitation (ChOP) using oligonucleotides corresponding to lncExACT1 sense sequence compared to oligonucleotides corresponding to lncExACT1 antisense sequence (as negative controls) pulled down DCHS2 promoter fragments detected by QRT-PCR corresponding to 500–1000 bp and 1000–1500 bp but not 0–500 bp upstream of DCHS2 (Fig. 5C). These data suggest lncExACT1 binds sequences between 500–1500 bp upstream of DCHS2 and positively regulates DCHS2 transcription. Since this occurs with exogenously expressed lncExACT1, it can occur in trans.
Figure 5. lncExACT1 works through DCHS2.
A. QRT-PCR measurement of DCHS2 in hearts from HF patients and controls (n=12/group), and in mice with HF or lncExACT1 overexpression, or exercise (Run) or lncExACT1 inhibition (Gap). B. Luciferase activity driven by DCHS2 promotor fragments in NRVMs with lncExACT1 OE or KD. C. QRT-PCR measurement with primers targeting different regions of DCHS2 promoter in complex from pulldown with probes targeting sense and antisense sequence of lncExACT1. D. Quantification of EdU and troponin double-positive cardiomyocytes by flow cytometry. E. Quantification of cardiomyocyte size in NRVMs treated with scrambled control or DCHS2 siRNA. F. QRT-PCR measurement of hypertrophy gene markers, n=3/group. G. Quantification of EdU positive cardiomyocytes by flow cytometry. H. Quantification of cardiomyocytes size after lncExACT1 OE and/or DCHS2 KD in NRVMs. *p<0.05, **p<0.01; in C and D, *p<0.05 vs. Control, #p<0.05 vs. lncExACT1 KD. A, C, D, and E by Student’s t test; B, F, G, and H by one-way analysis of variance (ANOVA) with post hoc Tukey. Data shown as mean±SEM.
In primary cardiomyocytes, small interfering RNA (siRNA) knockdown of DCHS2 (Fig.V.C in Data Supplement) induced an increase in cardiomyocyte proliferation as reflected by EdU incorporation (Fig. 5D). DCHS2 knockdown also increased cardiomyocyte size with a physiological gene expression pattern (Fig. 5E and Fig.V.D in Data Supplement). In contrast, while overexpression of DCHS2 using a CRISPR/dCas9 activation system also increased cardiomyocyte size, this was associated with a pathological hypertrophy gene expression pattern and reduced markers of cardiomyocyte proliferation (Fig.V.E–H in Data Supplement). Thus, similar to lncExACT1, DCHS2 inhibition induces physiological cardiomyocyte hypertrophy and proliferation while its overexpression induces pathological cardiomyocyte hypertrophy.
We then examined the interacting effects of DCHS2 and lncExACT1 in cardiomyocytes. DCHS2 knockdown prevented the pathological gene expression pattern otherwise seen with lncExACT1 overexpression (Fig. 5F) and DCHS2 expression blocked the physiological gene expression pattern seen with lncExACT1 knockdown (Fig.V.I in Data Supplement). Moreover, DCHS2 knockdown increased proliferation markers in cardiomyocytes overexpressing lncExACT1, while DCHS2 overexpression prevented the increase in cardiomyocyte proliferation induced by lncExACT1 inhibition (Fig. 5G and Fig.V.J in Data Supplement). Further, either knockdown or overexpression of DCHS2 prevented the increase in cardiomyocyte size seen with lncExACT1 overexpression (Fig. 5H) and knockdown (Fig.V.K in Data Supplement), respectively. Together these data suggest DCHS2 is necessary and sufficient for lncExACT1’s effects on cardiomyocyte growth and proliferation in vitro.
Although it has no known role in the heart, in other systems, DCHS2 modulates Hippo-Yap1 signaling24, a highly conserved regulator of size and proliferation in many organs including the heart25. Hippo activation ultimately induces phosphorylation and cytoplasmic sequestration of Yap1, which reduces proliferation, while its inhibition culminates in dephosphorylation and nuclear localization of Yap1 driving cell cycle progression26. Total nuclear Yap1 was increased while S127-phosphorylated cytoplasmic Yap1 (p-Yap1) was reduced in exercised hearts. The reverse was seen in failing hearts after TAC27, 28 (Fig.VI.A in Data Supplement). lncExACT1 inhibition in vivo also increased total nuclear Yap1 and reduced cytoplasmic p-Yap1. Conversely lncExACT1 overexpression increased cytoplasmic p-Yap1 and decreased total nuclear Yap1 (Fig.VI.B in Data Supplement). Similarly, either lncExACT1 or DCHS2 overexpression increased cytoplasmic p-Yap1 and reduced total nuclear Yap1 in primary cardiomyocytes (Fig.VI.C in Data Supplement). In contrast, lncExACT1 or DCHS2 inhibition in primary cardiomyocytes reduced cytoplasmic p-Yap1 and increased nuclear total Yap1 (Fig.VI.D in Data Supplement). These data suggest that physiological hypertrophy enhances while pathological hypertrophy inhibits Yap1 transcriptional activity likely through lncExACT1-DCHS2 signaling. Consistent with this model, knockdown of either lncExACT1 or DCHS2 in cardiomyocytes increased expression of multiple downstream targets of Yap126, 29, while lncExACT1 or DCHS2 expression had the opposite effect (Fig.VI.E–F in Data Supplement). Taken together these data implicate lncExACT1 and DCHS2 as novel regulators of cardiac Yap1, which likely contributes to exercise-induced heart growth and cardiomyogenesis.
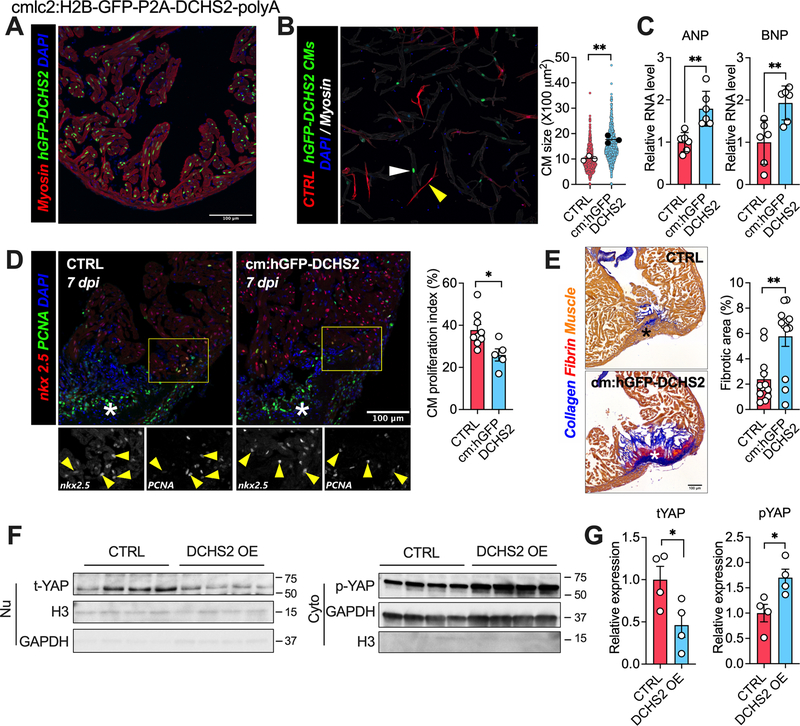
DCHS2 overexpression induces pathological cardiac hypertrophy and impairs cardiac regeneration in zebrafish
To examine DCHS2’s role in vivo, we turned to the zebrafish model, known for its remarkable ability to regenerate the heart after cardiac injury through cardiomyocyte proliferation30. We generated zebrafish constitutively expressing human DCHS2 in cardiomyocytes (Fig. 6A). Compared to controls, cardiac overexpression of DCHS2 increased cardiomyocyte size and expression of ANP and BNP, consistent with pathological cardiac hypertrophy (Fig. 6B–C). Seven days post cardiac cryoinjury (dpi), many proliferating cardiomyocytes (nxk2.5- and PCNA-positive) were evident in the injured area and border zone in wild-type animals. In contrast, significantly fewer were seen in DCHS2-expressing animals (Fig. 6D). We then asked whether the DCHS2-induced defect in cardiomyocyte proliferation would translate into long-term regeneration defects. At 90 dpi, all hearts (12 of 12) from wild-type animals had repaired the myocardial wall and showed only minimal residual fibrosis. In contrast, animals overexpressing DCHS2 developed substantially more fibrosis (Fig. 6E). These results indicate that DCHS2 impairs cardiac regeneration and promotes scarring after injury in zebrafish. Cardiac DCHS2 overexpression also reduced total nuclear Yap1 and increased cytoplasmic p-Yap1 (Fig. 6F–G). Altogether, these results indicate that cardiac overexpression of DCHS2 in zebrafish promotes pathological cardiac hypertrophy in unperturbed hearts, and impairs cardiomyocyte proliferation and regeneration, increasing fibrosis after cryoinjury. As in the mammalian studies, it seems likely that these effects are mediated through suppression of nuclear Yap1.
Figure 6. DCHS2 overexpression induces pathological cardiac hypertrophy and reduces regenerative capacity in zebrafish.
A. Representative image from apex region of an adult transgenic zebrafish heart immunostained for tropomyosin (Red), GFP (Green), and nuclei (DPAI, Blue). B. Representative image and quantification of cardiomyocytes size isolated from wild-type control (CTRL, white arrow) or DCHS2 overexpression (hGFP-DCHS2, yellow arrow) zebrafishes. C. QRT-PCR measurement of ANP and BNP in hearts from CTRL and hGFP-DCHS2 zebrafishes. D. Representative images and quantification of nkx2.5 and PCNA double positive cardiomyocytes in hearts at 7 days post injury (dpi) from CTRL and hGFP-DCHS2 zebrafish. E. Representative images and quantification of fibrosis in hearts at 60 days post injury (dpi) from CTRL and hGFP-DCHS2 zebrafish. F and G. Representative images and quantification of nuclear total Yap1 and cytoplasmic p-Yap1 protein expression in hearts from CTRL and hGFP-DCHS2 zebrafish. *p<0.05, **p<0.01 by Student’s t test. Data shown as mean±SEM.
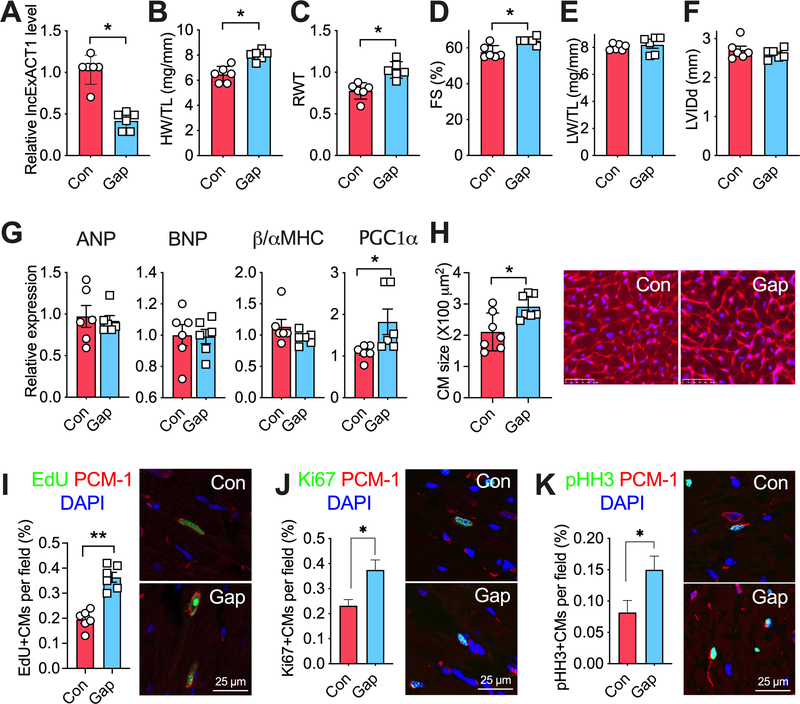
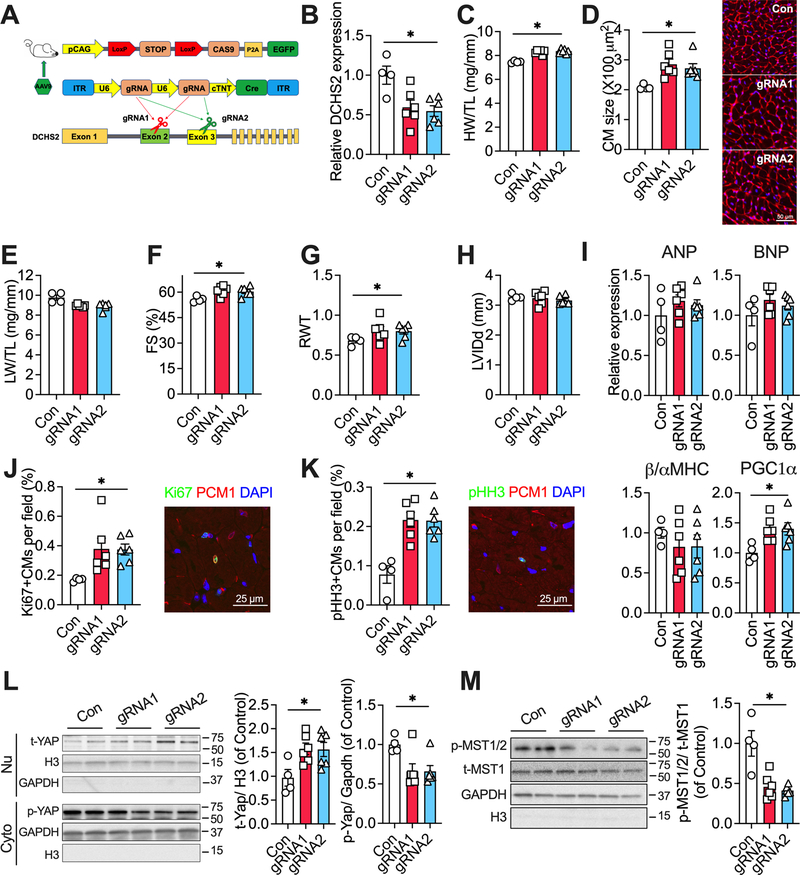
DCHS2 knock-down promotes physiological cardiac hypertrophy
To examine the effects of DCHS2 inhibition in vivo, we returned to the adult murine model which has limited cardiomyocyte proliferation at baseline. We injected Cas9 knock-in mice31, 32 with AAV9 vectors encoding either control guide RNAs (gRNAs), gRNAs targeting DCHS2 exon 2 (gRNA1), or exon 3 (gRNA2) along with a cardiac-specific troponin promoter-driven Cre expression cassette (Fig. 7A). Eight weeks after AAV injection, cardiac DCHS2 was reduced by ~50% in mice with gRNA1 or gRNA2 (Fig. 7B), which is comparable to the reduction seen in exercised hearts (Fig. 5F). Reduced cardiac DCHS2 led to increased HW/TL (Fig. 7C) and increased cardiomyocyte size (Fig. 7D) without affecting LW/TL (Fig. 7E). Reduced cardiac DCHS2 also improved cardiac function and increased relative wall thickness (Fig. 7F–G) without a significant change in chamber dimension (Fig. 7H). Reduced DCHS2 induced a gene expression pattern suggestive of physiological hypertrophy (Fig. 7I). These changes were qualitatively similar to those seen with comparable lncExACT1 knockdown but smaller quantitatively. Notably, DCHS2 knockdown increased cardiomyocyte proliferation as indicated by Ki67 and pHH3 in PCM1-positive cells (Fig. 7J–K) to a similar degree as seen with lncExACT1 knockdown (Fig. 3). Consistent with the in vitro and zebrafish studies, knockdown of DCHS2 increased nuclear total Yap1 but reduced cytoplasmic p-Yap1 expression in the heart (Fig. 7L). Knockdown of DCHS2 reduced cytoplasmic protein expression of p-MST1/2 (Thr183/Thr180) core regulatory proteins that work upstream of Yap1 in the Hippo pathway (Fig. 7M).33 Taken together, these data demonstrate that knockdown of DCHS2 in the adult mammalian heart is sufficient to induce physiological cardiac hypertrophy and evidence of cardiomyocyte proliferation. While the cardiomyocyte growth is modest, the increase in markers of proliferation is quantitatively similar to and thus likely sufficient to account for the that seen with lncExACT1 knockdown. The corresponding changes in nuclear Yap1 observed are likely to explain the proliferation changes seen with DCHS2 manipulation in both zebrafish and mammalian models.
Figure 7. DCHS2 knockdown promotes physiological cardiac hypertrophy.
A. Schematic of experimental strategy employed to knockdown DCHS2 in the hearts in vivo. B. QRT-PCR measurement of cardiac DCHS2 in Cas9 knockin mice with injection of AAV9 carrying gRNA1 or gRNA2. C. Heart weight (HW) relative to tibial length (TL). D. Quantification of cardiomyocyte area from wheat germ agglutinin (WGA)-stained heart sections. E. Lung weight (LW) relative to tibial length (TL). F. Fractional shortening (FS). G. Relative wall thickness (RWT). H. Left ventricular end diastolic internal dimension (LVIDd). I. QRT-PCR measurement of hypertrophy gene markers. J. Quantification of Ki67. K. Quantification of pHH3, PCM1 double-positive cardiomyocytes in stained heart sections. L and M. Representative images and quantification of nuclear total Yap1, and cytoplasmic p-Yap1, p-MST1/2, and total MST1 (t-MST1) protein expressions in hearts from mice injected with AAV9 carrying control, or gRNA1, or gRNA2. *p<0.05 by one-way analysis of variance (ANOVA) with post hoc Tukey. Data shown as mean±SEM.
Discussion
Understanding how exercise promotes cardiac health and whether the responsible mechanisms can be feasibly targeted has important fundamental and clinical implications. The data presented here identify a set of cardiac lncRNAs (lncExACTs) that are dynamically regulated by exercise and play an important role in determining the outcome of cardiac growth. A small number of the lncExACTs identified were also altered in pathological hypertrophy or HF. In every case, the change in exercise was opposite to that observed in disease models, underscoring the distinct nature of these responses despite superficial similarities. Among these, lncExACT1 was uniquely downregulated in exercise and highly conserved across species. lncExACT1 expression was sufficient to induce pathological hypertrophy and HF. Inhibition of lncExACT1 was sufficient to induce multiple exercise-related cardiac phenotypes including physiological hypertrophy, improved cardiac function, protection against HF and fibrosis, and signs of cardiomyocyte proliferation.
Surprisingly both lncExACT1 overexpression and inhibition induced cardiac hypertrophy. However, several lines of evidence suggest that lncExACT1 overexpression produced pathological growth in contrast to the physiological growth induced by inhibition. First, lncExACT1 gain- and loss-of-function induced distinct patterns in expression of the sentinel markers of pathological vs physiological growth4, 5. Second, lncExACT1 inhibition increased markers of cardiomyocyte proliferation, a hallmark of exercise-induced but not pathological growth, both at baseline and in disease models. lncExACT1 overexpression did not affect markers of cardiomyocyte proliferation in vivo. Finally, and perhaps most important, lncExACT1 overexpression culminated in cardiac dysfunction and HF while inhibition actually improved cardiac function modestly at baseline and more significantly after pathological stress. Taken together, these data strongly support the model that the cardiomyocyte growth induced by lncExACT1 overexpression and inhibition are fundamentally different, representing pathological and physiological growth, respectively. Of note, the expression level of lncRNAs is low compared to protein coding genes. Prior work suggests that for lncRNA cis effects an abundance of 1–10 molecules per cell can be sufficient34, while for trans action, an abundance of 10–1000 has been reported35. We found there are 35.59±3.60 copies of lncExACT1 per cardiomyocyte at baseline, which increases by 4–8-fold in HF and decreases 2–3-fold in exercise. These changes could plausibly account for its regulatory effects on DCHS2 and miR-222. To our knowledge, this is a unique example of one molecule whose expression toggles the heart between pathological and physiological growth with very different functional outcomes and suggests targeting lncExACT1 may hold therapeutic promise.
The adult mammalian heart has a very limited capacity for cardiomyogenesis, estimated at ~1%/year in both humans36 and mice37. This limitation has important clinical consequences since heart disease is often associated with cardiomyocyte loss38. To date, exercise is the only physiological stimulus demonstrated to increase cardiomyogenesis in the adult mammalian heart10. While prior work demonstrated that miR-222 is necessary for this effect, miR-222 alone was not sufficient to recapitulate cardiac exercise phenotypes, including cardiomyogenesis10, 11. Collectively, the studies presented here implicate a novel pathway including the lncRNA, lncExACT1, and its effector, DCHS2, as candidate regulators of exercise-induced cardiomyogenesis. The zebrafish studies demonstrate a role for DCHS2 in regulating cardiomyocyte proliferation and regeneration with important consequences for healing and scar formation. These findings resonate with the murine data where lncExACT1 inhibition increased markers of cardiomyocyte proliferation after TAC and in the border zone after IRI, with a corresponding reduction in late scar formation and improved cardiac function. However, the murine studies presented rely on surrogate markers of DNA synthesis which cannot conclusively establish cardiomyogenesis or its role in the functional benefits seen.
The observed effects on fibrosis are consistent with prior work demonstrating that exercise itself reduces cardiac fibrosis after aortic constriction or ischemic injury39. The ~2-fold reduction in fibrosis seen with lncExACT1 knockdown after TAC or IRI could be secondary to effects in cardiomyocytes, including increased cardiomyocyte survival and proliferation leading to less replacement fibrosis. This model is supported by the zebrafish studies in which cardiomyocyte-specific expression of DCHS2 reduced cardiomyocyte proliferation impairing cardiac regeneration and culminating in greater fibrosis (Fig.6). This pathway may also have direct effects on fibroblast proliferation opposite to its effects in cardiomyocytes, as suggested in our in vitro studies (Fig.III in Data Supplement). Although lncExACT1 knockdown was moderately less efficient in fibroblasts compared to cardiomyocytes, this seems unlikely to account for the qualitatively opposite effect on proliferation. It seems likely a combination of all these effects contributes to the reduced fibrosis and cardiac benefits of lncExACT1 or DCHS2 inhibition.
These studies also implicate DCHS2, not previously known to have a role in the heart, as an important modulator of cardiac growth and cardiomyocyte proliferation as well as a downstream effector of lncExACT1. lncExACT1 binds the promoter of DCHS2, positively regulating its transcription. DCHS2 has been demonstrated in other systems to work with FAT as a ligand-receptor system in regulating cell proliferation through Hippo/Yap signaling24, 40. Consistent with this, we found that exercise, as well as inhibition of cardiac lncExACT1 or DCHS2, increased nuclear Yap1 and expression of its downstream targets. Our results suggest that DCHS2 may regulate Yap1, at least in part, through inhibiting MST1/2 phosphorylation, although the precise mechanism remains to be elucidated. To our knowledge, these are also the first studies to implicate Hippo/Yap1 signaling, a recognized regulator of cardiomyogenesis in the adult heart25, in exercise-induced cardiomyogenesis10.
The role of Hippo/Yap1 signaling in the cardiac hypertrophy observed is less clear. Prior studies demonstrated that postnatal activation of cardiac Yap1 did not increase cardiac or cardiomyocyte size41, 42, suggesting Yap1 activation is not sufficient to induce physiological cardiac growth. In contrast cardiac deletion of Yap1 reduced hypertrophy in response to pressure overload43. Taken together, the previous work and our data suggest the effects observed on cardiac hypertrophy are likely mediated by Yap1-independent pathways. In vitro studies suggest lncExACT1 binding of miR-222, which we had previously shown is necessary for cardiac growth11 and exercise-induced cardiomyogenesis10 in vivo, contributes to lncExACT1’s effects in physiological hypertrophy (Fig.IV in Data Supplement). In vitro studies also suggest that calcineurin signaling may contribute to the pathological hypertrophy and/or HF seen with lncExACT1 overexpression. We think it is also likely that other downstream contributors remain to be uncovered and will be a focal point for future work.
lncRNAs have well-documented roles in cardiac development,44, 45 pathological hypertrophy,46, 47 and HF48, 49. However, less is known about their roles in exercise. A recent report by Gao and colleagues50 identified a lncRNA, CPhar, induced by exercise in murine hearts. CPhar was not one of the lncRNAs detected in our screen, perhaps reflecting differences in the time-points (3 vs 8 weeks) and exercise models (swim vs voluntary running). Interestingly, the prior study demonstrated that CPhar was necessary for exercise-induced hypertrophy and, when expressed prior to ischemic injury, significantly mitigated cardiac dysfunction50.
We believe the present work adds to this literature in important conceptual and practical ways. First, by comprehensively comparing cardiac lncRNAs dynamically regulated in both exercise and pathological states, we were able to identify a subset of lncRNAs altered in both (but in opposing ways), which speaks to the fundamental differences between these states. This lncRNA set is likely enriched for functional candidates contributing to both states and worthy of evaluation as therapeutic targets. Second, mimicking the change in lncExACT1 seen in exercise is sufficient to recapitulate many exercise-induced cardiac phenotypes in vivo, including evidence of cardiomyogenesis whereas it appears CPhar expression (similar to miR-222) was not sufficient to reproduce these general phenotypes or cardiomyogenesis in vivo50. Pathways sufficient to reproduce the benefits of exercise, including cardiomyogenesis, are particularly unusual and valuable.
lncExACT1 has other strategic advantages as a therapeutic target. It is highly (~70%) conserved between mice and humans, and altered similarly in human and experimental HF; while most lncRNAs are not well conserved. Importantly, benefits are observed with lncExACT1 inhibition rather than overexpression. Multiple modified nucleic acid antisense drugs have been approved by the FDA, including some utilizing the locked nucleic acid (LNA) chemistry employed here51 and both large animal and early clinical studies support the feasibility of using LNA-antisense delivery to inhibit noncoding cardiac RNAs52, 53. These practical advantages enabled us to demonstrate a substantive reduction in cardiac scar and improvement in function even when inhibitor was delivered after reperfusion, a clinically relevant timeframe. In contrast, targets requiring anticipatory overexpression can be challenging to translate clinically given the generally unanticipated nature of ischemic injury. Nevertheless, both studies reinforce the message that much can be learned from lncRNAs mediating the benefits of exercise.
In summary, these studies identify the lncExACT1-DCHS2 pathway as a crucial and previously unrecognized regulator of both physiological and pathological cardiomyocyte growth, integrating the dynamic tension between these states, and influencing clinically relevant outcomes in each. Targeting this pathway therapeutically warrants further investigation in a range of cardiac diseases.
Supplementary Material
Clinical Perspective.
What is new?
lncExACT1 is a conserved long noncoding RNA that decreases in exercised hearts but increases in failing hearts from animals and people
lncExACT1 overexpression induces pathological hypertrophy and cardiac dysfunction, while its inhibition induces physiological growth and protects against dysfunction and fibrosis.
What are the clinical implications?
lncExACT1 inhibition reduces cardiomyocyte loss and scar formation while improving cardiac function after pathological stress including ischemic injury and pressure-overload.
Interventions including modified nucleic acid antisense drugs, which have been approved by the FDA for other indications, can effectively inhibit cardiac lncExACT1 to mitigate cardiac dysfunction and heart failure in animal models, and warrant further investigation as a therapeutic strategy.
Acknowledgements
We thank Drs. Ling Li (Cardiovascular Research Center, MGH) for technical support and James Rhee (Department of Anesthesiology, MGH) for sharing RILPL1-Flag tag plasmid, and Yoshiko Iwamoto (Center for Systems Biology, MGH) for helping with sample staining.
Sources of Funding
This research is supported by the NIH (R01AG061034, R35HL155318 to A.R.), AHA (20CDA35310184 to H.L., 19CDA34660207 to J.M.G., AHA 16SFRN31720000 to A.R., 19POST34381027 to G.L.), NIH/NCATS Colorado CTSA (UL1 TR002535 to A.V.A and M.B.), Alan Hassenfeld Research Scholar Award to J.M.G., Sarnoff Cardiovascular Research Foundation Fellowship to L.E.T., and reflects a collaboration across AHA SFRN Heart Failure Centers.
Nonstandard Abbreviations and Acronyms
- MHC
myosin heavy chain
- MIMS
multi-isotope imaging mass spectrometry
- lncRNA
long noncoding RNA
- lncExACT
long noncoding exercise-associated cardiac transcript
- DCHS2
dachsous cadherin-related 2
- TAC
transvers aortic constriction
- IGF-1
insulin-like growth factor 1
- ddPCR
droplet digital PCR
- SVT
supraventricular tachycardia
- AAV
adeno-associated virus
- IRI
ischemia reperfusion injury
- ceRNA
competitive endogenous RNAs
Footnotes
Disclosures
H.L. and A.R. are inventors on a pending patent (63/058,268: Inhibition of lncExACT1 to Treat Heart Disease) submitted by MGH. Other authors declare no competing interest.
Supplementary Materials
References
- 1.Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, et al. ; American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation. 2020;141:e139–e596. [DOI] [PubMed] [Google Scholar]
- 2.Pandey A, LaMonte M, Klein L, Ayers C, Psaty BM, Eaton CB, Allen NB, de Lemos JA, Carnethon M, Greenland P, et al. Relationship Between Physical Activity, Body Mass Index, and Risk of Heart Failure. J Am Coll Cardiol. 2017;69:1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang B, Xu M, Li W, Li X, Zheng Q, Niu X. Aerobic exercise protects against pressure overload-induced cardiac dysfunction and hypertrophy via beta3-AR-nNOS-NO activation. PLoS One. 2017;12:e0179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iemitsu M, Miyauchi T, Maeda S, Sakai S, Kobayashi T, Fujii N, Miyazaki H, Matsuda M, Yamaguchi I. Physiological and pathological cardiac hypertrophy induce different molecular phenotypes in the rat. Am J Physiol Regul Integr Comp Physiol. 2001;281:R2029–2036. [DOI] [PubMed] [Google Scholar]
- 5.Bostrom P, Mann N, Wu J, Quintero PA, Plovie ER, Panakova D, Gupta RK, Xiao C, MacRae CA, Rosenzweig A, et al. C/EBPbeta controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell. 2010;143:1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdul-Ghani M, Suen C, Jiang B, Deng Y, Weldrick JJ, Putinski C, Brunette S, Fernando P, Lee TT, Flynn P, et al. Cardiotrophin 1 stimulates beneficial myogenic and vascular remodeling of the heart. Cell Res. 2017;27:1195–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, Akazawa H, Tateno K, Kayama Y, Harada M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. [DOI] [PubMed] [Google Scholar]
- 8.Narula J, Haider N, Virmani R, DiSalvo TG, Kolodgie DF, Hajjar RJ, Schmidt U, Semigran MJ, Dec GW, Khaw B-A. Apoptosis in myocytes in end-stage heart failure. N Engl J Med. 1996;335:1182–1189. [DOI] [PubMed] [Google Scholar]
- 9.Eschenhagen T, Bolli R, Braun T, Field LJ, Fleischmann BK, Frisen J, Giacca M, Hare JM, Houser S, Lee RT, et al. Cardiomyocyte Regeneration: A Consensus Statement. Circulation. 2017;136:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vujic A, Lerchenmuller C, Wu TD, Guillermier C, Rabolli CP, Gonzalez E, Senyo SE, Liu X, Guerquin-Kern JL, Steinhauser ML, et al. Exercise induces new cardiomyocyte generation in the adult mammalian heart. Nat Commun. 2018;9:1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, Xiao C, Bezzerides V, Bostrom P, Che L, Zhang C, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21:584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMullen JR, Shioi T, Zhang L, Tarnavski O, Sherwood MC, Kang PM, Izumo S. Phosphoinositide 3-kinase(p110alpha) plays a critical role for the induction of physiological, but not pathological, cardiac hypertrophy. Proc Natl Acad Sci USA. 2003;100:12355–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeBosch B, Treskov I, Lupu TS, Weinheimer C, Kovacs A, Courtois M, Muslin AJ. Akt1 is required for physiological cardiac growth. Circulation. 2006;113:2097–2104. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Wende AR, Sena S, Theobald HA, Soto J, Sloan C, Wayment BE, Litwin SE, Holzenberger M, LeRoith D et al. Insulin-like growth factor I receptor signaling is required for exercise-induced cardiac hypertrophy. Mol Endocrinol. 2008;22:2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue TL, Gu JL, Wang C, Reith AD, Lee JC, Mirabile RC, Kreutz R, Wang Y, Maleeff B, Parsons AA et al. Extracellular signal-regulated kinase plays an essential role in hypertrophic agonists, endothelin-1 and phenylephrine-induced cardiomyocyte hypertrophy. J Biol Chem. 2000;275:37895–37901. [DOI] [PubMed] [Google Scholar]
- 16.De Windt LJ, Lim HW, Bueno OF, Liang Q, Delling U, Braz JC, Glascock BJ, Kimball TF, del Monte F, Hajjar RJ et al. Targeted inhibition of calcineurin attenuates cardiac hypertrophy in vivo. Proc Natl Acad Sci USA. 2001;98:3322–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See K, Tan WLW, Lim EH, Tiang Z, Lee LT, Li PYQ, Luu TDA, Ackers-Johnson M, Foo RS. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei X, Liu X, Rosenzweig A. What do we know about the cardiac benefits of exercise? Trends Cardiovasc Med. 2015;25:529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campos JC, Queliconi BB, Bozi LHM, Bechara LRG, Dourado PMM, Andres AM, Jannig PR, Gomes KMS, Zambelli VO, Rocha-Resende C, et al. Exercise reestablishes autophagic flux and mitochondrial quality control in heart failure. Autophagy. 2017;13:1304–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh PCH, Segers VFM, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13:970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park M, Vatner SF, Yan L, Gao S, Yoon S, Lee GJ, Xie LH, Kitsis RN, Vatner DE. Novel mechanisms for caspase inhibition protecting cardiac function with chronic pressure overload. Basic Res Cardiol. 2013;108:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, Dorn GW 2nd, Thum T, Heymans S; Cardiolinc Network. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol. 2015;12:415–425. [DOI] [PubMed] [Google Scholar]
- 24.Bagherie-Lachidan M, Reginensi A, Pan Q, Zaveri HP, Scott DA, Blencowe BJ, Helmbacher F, McNeill H. Stromal Fat4 acts non-autonomously with Dchs1/2 to restrict the nephron progenitor pool. Development. 2015;142:2564–2573. [DOI] [PubMed] [Google Scholar]
- 25.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL,Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Liu S, Heallen T, Martin JF. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol. 2018;15:672–684. [DOI] [PubMed] [Google Scholar]
- 27.Morikawa Y, Heallen T, Leach J, Xiao Y, Martin JF. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature. 2017;547:227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda S, Mizushima W, Sciarretta S, Abdellatif M, Zhai P, Mukai R, Fefelova N, Oka SI, Nakamura M, Del Re DP, et al. Hippo Deficiency Leads to Cardiac Dysfunction Accompanied by Cardiomyocyte Dedifferentiation During Pressure Overload. Circ Res. 2019;124:292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Totaro A, Panciera T, Piccolo S. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol. 2018;20:888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, Lu YW, Lin Z, Huang ZP, Liu J, Wang Y, Seok HY, Hu X, Ma Q, Li K, et al. Intercalated disc protein Xinbeta is required for Hippo-YAP signaling in the heart. Nat Commun. 2020;11:4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moya IM, Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol. 2019;20:211–226. [DOI] [PubMed] [Google Scholar]
- 34.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, Stern-Ginossar N, Ulitsky I. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Comm. 2016;7:12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foo RS, Mani K, Kitsis RN. Death begets failure in the heart. J Clin Invest. 2005;115:565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Wan W, Powers AS, Li J, Ji LL, Lao S, Wilson B, Erikson JM, Zhang JQ. Effects of exercise training on cardiac function and myocardial remodeling in post myocardial infarction rats. J Mol Cell Cardiol. 2008;44:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma P, McNeill H. Fat and Dachsous cadherins. Prog Mol Biol Transl Sci. 2013;116:215–235. [DOI] [PubMed] [Google Scholar]
- 41.Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, Yau AL, Buck JN, Gouin KA, van Gorp PR, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 2014;115:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A. 2012;109:2394–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byun J, Del Re DP, Zhai P, Ikeda S, Shirakabe A, Mizushima W, Miyamoto S, Brown JH, Sadoshima J. Yes-associated protein (YAP) mediates adaptive cardiac hypertrophy in response to pressure overload. J Biol Chem. 2019;294:3603–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizki G, Boyer LA. Lncing epigenetic control of transcription to cardiovascular development and disease. Circ Res. 2015;117:192–206. [DOI] [PubMed] [Google Scholar]
- 45.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sallam T, Sandhu J, Tontonoz P. Long Noncoding RNA Discovery in Cardiovascular Disease: Decoding Form to Function. Circ Res. 2018;122:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viereck J, Thum T. Long Noncoding RNAs in Pathological Cardiac Remodeling. Circ Res. 2017;120:262–264. [DOI] [PubMed] [Google Scholar]
- 50.Gao R, Wang L, Bei Y, Wu X, Wang J, Zhou Q, Tao L, Das S, Li X, Xiao J. LncRNA CPhar Induces Cardiac Physiological Hypertrophy and Promotes Functional Recovery After Myocardial Ischemia-Reperfusion Injury. Circulation. 2021;144:303–317. [DOI] [PubMed] [Google Scholar]
- 51.Dhuri K, Bechtold C, Quijano E, Pham H, Gupta A, Vikram A, Bahal R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J Clin Med. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foinquinos A, Batkai S, Genschel C, Viereck J, Rump S, Gyongyosi M, Traxler D, Riesenhuber M, Spannbauer A, Lukovic D, et al. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat Commun. 2020;11:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, Rode L, Weigt H, Genschel C, Lorch U, et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J. 2021;42:178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, Schier AF. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development. 2013;140:2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roh JD, Hobson R, Chaudhari V, Quintero P, Yeri A, Benson M, Xiao C, Zlotoff D, Bezzerides V, Houstis N, et al. Activin type II receptor signaling in cardiac aging and heart failure. Sci Transl Med. 2019;11:eaau8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016;8:326ra22. [DOI] [PubMed] [Google Scholar]
- 57.Bezzerides VJ, Platt C, Lerchenmuller C, Paruchuri K, Oh NL, Xiao C, Cao Y, Mann N, Spiegelman BM, Rosenzweig A. CITED4 induces physiologic hypertrophy and promotes functional recovery after ischemic injury. JCI Insight. 2016;1:e85904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Yao W, Liu Z, Xu A, Huang Y, Ma XL, Irwin MG, Xia Z. Hyperglycemia Abrogates Ischemic Postconditioning Cardioprotection by Impairing AdipoR1/Caveolin-3/STAT3 Signaling in Diabetic Rats. Diabetes. 2016;65:942–955. [DOI] [PubMed] [Google Scholar]
- 60.Matsui T, Tao J, del Monte F, Lee KH, Li L, Picard M, Force TL, Franke TF, Hajjar RJ, Rosenzweig A. Akt activation preserves cardiac function and prevents injury after transient cardiac ischemia in vivo. Circulation. 2001;104:330–335. [DOI] [PubMed] [Google Scholar]
- 61.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FC, Bisaria A, Betzig E, Meyer T. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol. 2016;18:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pleger ST, Shan C, Ksienzyk J, Bekeredjian R, Boekstegers P, Hinkel R, Schinkel S, Leuchs B, Ludwig J, Qiu G, et al. Cardiac AAV9-S100A1 gene therapy rescues post-ischemic heart failure in a preclinical large animal model. Sci Transl Med. 2011;3:92ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y, VanDusen NJ, Zhang L, Gu W, Sethi I, Guatimosim S, Ma Q, Jardin BD, Ai Y, Zhang D, et al. Analysis of Cardiac Myocyte Maturation Using CASAAV, a Platform for Rapid Dissection of Cardiac Myocyte Gene Function In Vivo. Circ Res. 2017;120:1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Torres M, Becquet D, Guillen S, Boyer B, Moreno M, Blanchard MP, Franc JL, Francois-Bellan AM. RNA Pull-down Procedure to Identify RNA Targets of a Long Non-coding RNA. J Vis Exp. 2018; 57379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossi MN, Maione R. Identification of Chromatin Binding Sites for Long Noncoding RNAs by Chromatin Oligo-Affinity Precipitation (ChOP). Methods Mol Biol. 2020;2161:17–28. [DOI] [PubMed] [Google Scholar]
- 70.Gonzalez-Rosa JM, Mercader N. Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat Protoc. 2012;7:782–788. [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalez-Rosa JM, Sharpe M, Field D, Soonpaa MH, Field LJ, Burns CE, Burns CG. Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev Cell. 2018;44:433–446 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.