Abstract
Ixeris sonchifolia extract injection, a Chinese medicine preparation named as Kudiezi injection (KDZI) in China, has been widely used for the treatment of cardiovascular diseases (CVDs) in recent years. Owing to the component complexity of the preparation, the study on the effect mechanism of the herbal medicine against CVDs is a big challenge. In this research, HPLC-Q-TOF-MS was used to analyze the constituents of the preparation, disclosing that the KDZI mainly consists of 10 ingredients, namely 3-caffeoylquinic acid (KDZI-1), 4-caffeoylquinic acid (KDZI-2), 5-caffeoylquinic acid (KDZI-3), apigenin-7-O-β-d-glucuronide (KDZI-4), caffeic acid (KDZI-5), chicoric acid (KDZI-6), caftaric acid (KDZI-7), luteolin-7-O-β-d-gentiobioside (KDZI-8), luteolin-7-O-β-d-glucopyranoside (KDZI-9) and luteolin-7-O-β-d-glucuronide (KDZI-10). Afterwards, target fishing and an integrated systems pharmacology approach combined with molecular docking (Sybyl 1.3 and AutoDock Vina) were adopted to predict the potential targets and pathways for the main ingredients in KDZI. As results, 39 protein targets and 9 KEGG pathways, possessing high relevance to the therapeutic effects of the ingredients of KDZI against CVDs, were screened out reasonably. The integrated pharmacology analysis suggested that KDZI could exert its therapeutic effects against CVDs possibly via multi-targets including EGFR, MAPK10, and SRC and multi-pathways referring to MAPK, focal adhesion, complement and coagulation cascades, etc. This research provides insights into understanding the comprehensive therapeutic effect and mechanism of the KDZI on CVDs.
Ixeris sonchifolia extract injection, a Chinese medicine preparation named as Kudiezi injection (KDZI) in China, has been widely used for the treatment of cardiovascular diseases (CVDs) in recent years.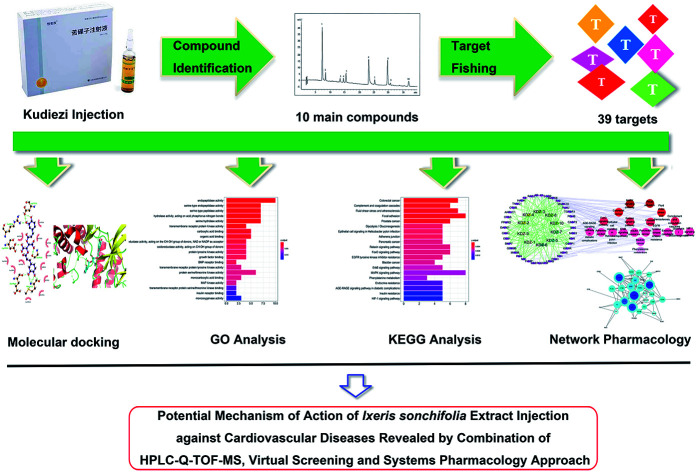
1. Introduction
Cardiovascular diseases (CVDs) refer to the heart, brain and tissue ischemia or hemorrhagic diseases mainly caused by hyperlipidemia, blood viscosity, atherosclerosis, hypertension, etc.1 Currently, CVDs are still the leading cause of morbidity and mortality globally.2 More deaths have been recorded due to CVDs than from any other cause, and possibly 23.6 million deaths per year by 2030 is reported by the World Heart Federation.3 In 2018, there were 290 million patients suffering from CVDs in China.4 In China, CVDs are believed to be the number one killer in the future. Hence, it is significant to explore new therapeutic drugs or methods to prevent and treat CVDs.
Herbal medicines have been gradually recognized worldwide as safe and effective therapeutics with fewer side effects.5–11 Some herbal medicine injections produced by extracting and purifying active ingredients from medicinal plants according to modern scientific and technological theories have often been used to treat some emergency and severe diseases, such as CVDs and stock in clinic in China. Ixeris sonchifolia extract injection, a Chinese medicine preparation named as Kudiezi injection (KDZI) in China, is extracted from the herb Ixeris sonchifolia (Bunge) Hance and extensively applied to treat CVDs, such as ischemic stroke, myocardial infarction, etc.12 Studies have shown that KDZI mainly contains flavonoids, adenosine, sesquiterpene lactones and triterpene saponins.13 However, the therapeutic effect of KDZI against CVDs remains unclear due to complexity of its components.
In the recent years, owing to the convenient and powerful properties on exploring the inner relationship among multiple ingredients, multiple targets and multiple pathways for a complex drug system, the systems pharmacology study has become a popular trend on dissecting the mechanism of action compound formulas, e.g., traditional Chinese medicines, herbal medicines or their preparation products.14–19 Also, systems pharmacology was considered to provide valuable clues on identifying the active ingredients from a complex herbal formula.17–19 However, up to date, there is still lack of the comprehensive study on dissecting the effect mechanisms of KDZI based on the systems pharmacology methods.
In this study, an integrated approach was established to unveil the pharmacological mechanisms of the main constitutions in KDZI from the whole perspective of system biology. First, the main constituents in KDZI were identified by HPLC-Q-TOF-MS. Then, molecular docking simulations, KEGG (Kyoto Encyclopedia of Genes and Genomes) enriched pathway analysis, GO (Gene Ontology) analysis and network analysis were further employed to explore the anti-CVD mechanism of KDZI.
2. Materials and methods
2.1. HPLC-Q-TOF-MS analysis
For the HPLC-Q TOF-MS analysis, an Agilent 1260 HPLC instrument and an Agilent 6520 quadrupole time-of-flight tandem mass spectrometer were used in this study. The separation was finally performed on an Agilent ZORBAX SB-C18 column (4.6 mm × 150 mm, 3.5 μm) at 30 °C at a flow rate of 0.5 mL min−1. The mobile phase consisted of 0.5% glacial acetic acid in water (A) and acetonitrile (B) with the following gradient elution: 0–15 min, 4–11% B; 15–20 min, 11–14% B; 20–40 min, 14–19% B; 40–41 min, 19–95% B. The injection volume was 5 μL.
The acquisition parameters for Q-TOF mass spectra were as follows: drying gas (N2) flow rate; 10 L min−1, drying gas temperature, 350 °C; atomizing gas flow, 40 psi; electricity spray voltage 3500 V, fragmentor voltage 145 V. The sample was analyzed in both positive and negative ion modes for full scan (ESI†), and the scan mass range was recorded from m/z 100 to 1000.
2.2. Target predicting
First, three dimensional molecule structures of the identified compounds were constructed using a molecular docking software Sybyl-X. Subsequently, an online target fishing sever, PharmMapper (http://lilab.ecust.edu.cn/pharmmapper), which uses a pharmacophore mapping approach probing potential drug target, was used to predict the target proteins of these compounds.20–23 The predicted results with fit score, related gene ID, and the correlated diseases, etc. were documented using an excel table. Next, the results were filtered by a comprehensive consideration of norm fit (≥0.7) and the definite relationship of the target proteins with cardiovascular diseases (searching PubMed, Web of science and Genecards databases).
2.3. Molecular docking
Molecular docking, an important means of modern drug screening, was used to analyze the binding ability of compounds to target proteins.24 First, the crystal structures of the target proteins were downloaded from the PDB database (https://www.rcsb.org/) and embellished through the Sybyl-X,25 including removing waters and ligands, adding H atoms, optimizing and patching amino acids. The docking scores, including total scores (preferable ≥ 6) and CScores (preferable ≥ 4), were the important evaluation indices for predicting the affinity of a target protein to an ingredient. Besides, Autodock Vina,26 an open-source program for molecular docking and virtual screening, was also used in this study to access the binding affinity of each compound/protein complex. It was reported that this program had a high prediction performance when compared to experimental data.27 With the application of the two molecule docking methods, the total-score and affinity values (kcal mol−1) were used to evaluate the interaction between ingredients and targets, respectively. Moreover, the better docked model was analyzed using PyMOL28 and Ligplus.29
2.4. GO and KEGG pathway enrichment analyses
GO and KEGG analyses were adopted for the 39 potential targets, which were performed by the clusterProfiler package in R (version 3.5.1) to address the functional annotation (p < 0.01).30
2.5. Network construction
The 39 potential targets were inputted into String database (https://string-db.org/cgi/input.pl) to build a PPI network and illustrated the inter-protein interaction (confidence scores > 0.4). Besides, compounds, candidate targets and pathways for KDZI were used to construct the compound-target-pathway network visualized by the Cytoscape software.31
3. Results and discussion
3.1. Identification of the main chemical constituents in KDZI
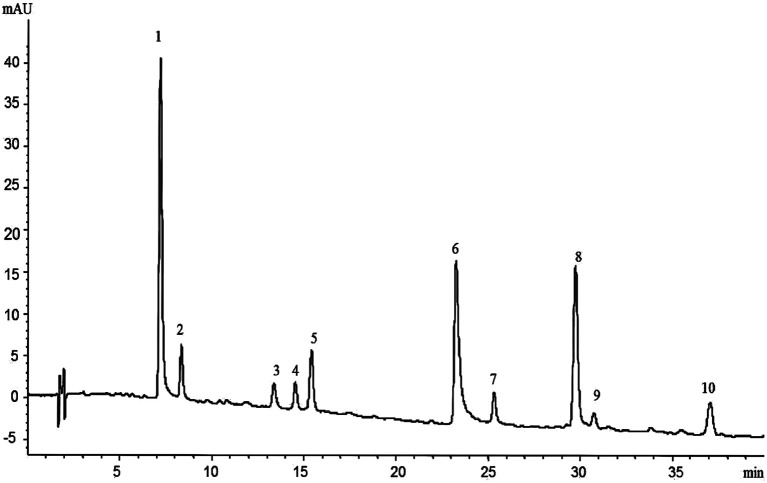
The HPLC-Q-TOF-MS analysis of the KDZI was performed in both positive and negative ionization modes. The HPLC chromatogram is shown in Fig. 1, and the MS data are listed in Table 1. According to Fig. 1, 10 main compounds were identified, including flavonoids, esters, and organic acids, with their standard references. The chemical structures are listed in Table 2.
Fig. 1. HPLC chromatogram of KDZI.
Characterization of compounds in KDZI injection by HPLC-Q TOF-MS.
| No. | t R (min) | Observed mass | Calculated mass | Error (ppm) | Formula | Identification |
|---|---|---|---|---|---|---|
| 1 | 7.376 | 311.0397 | 311.0409 | 3.83 | C13H12O9 | Caftaric acid (KDZI-7) |
| 2 | 8.518 | 353.0855 | 353.0878 | 6.62 | C16H18O9 | 5-Caffeoylquinic acid (KDZI-3) |
| 3 | 13.342 | 353.0866 | 353.0878 | 3.52 | C16H18O9 | 3-Caffeoylquinic acid (KDZI-1) |
| 4 | 14.870 | 353.0869 | 353.0878 | 2.62 | C16H18O9 | 4-Caffeoylquinic acid (KDZI-2) |
| 5 | 15.384 | 179.0347 | 179.035 | 1.67 | C9H8O4 | Caffeic acid (KDZI-5) |
| 6 | 23.586 | 473.0692 | 473.0692 | 7.20 | C22H18O12 | Chicoric acid (KDZI-6) |
| 7 | 25.580 | 609.1432 | 609.1461 | 4.90 | C27H30O16 | Luteolin-7-O-β-d-gentiobioside (KDZI-8) |
| 8 | 29.986 | 461.0726 | 461.0725 | −0.02 | C21H18O12 | Luteolin-7-O-β-d-glucuronide (KDZI-10) |
| 9 | 30.742 | 447.0918 | 447.0933 | 3.37 | C21H20O11 | Luteolin-7-O-β-d-glucopyranoside (KDZI-9) |
| 10 | 37.223 | 445.0761 | 445.0776 | 3.55 | C21H18O11 | Apigenin-7-O-β-d-glucuronide (KDZI-4) |
Main ingredients and related parameters of KDZI.
| No. | Name | Molecular formula | Molecular weight | Structure |
|---|---|---|---|---|
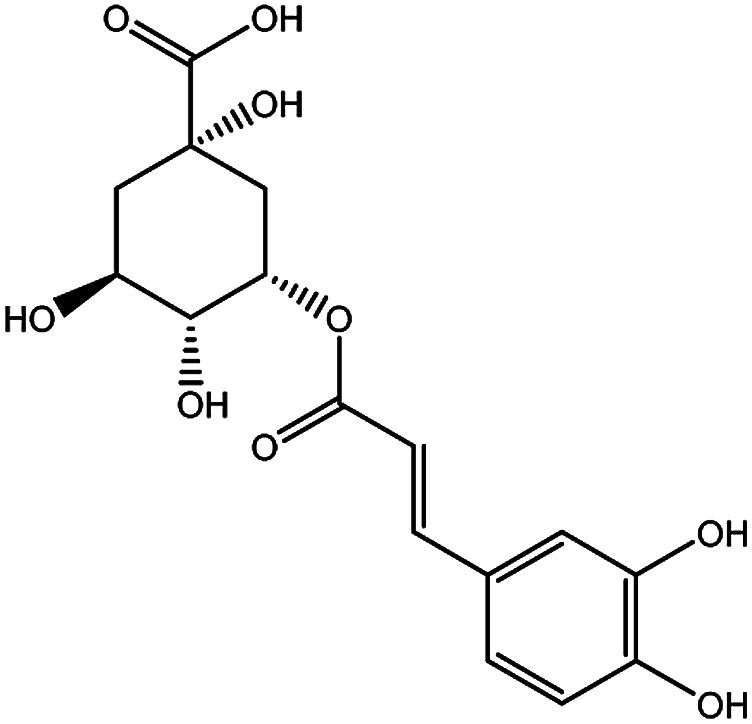
| KDZI-1 | 3-Caffeoylquinixacid | C16H18O9 | 354.31 |
|
| KDZI-2 | 4-Caffeoylquinic acid | C16H18O9 | 354.31 |
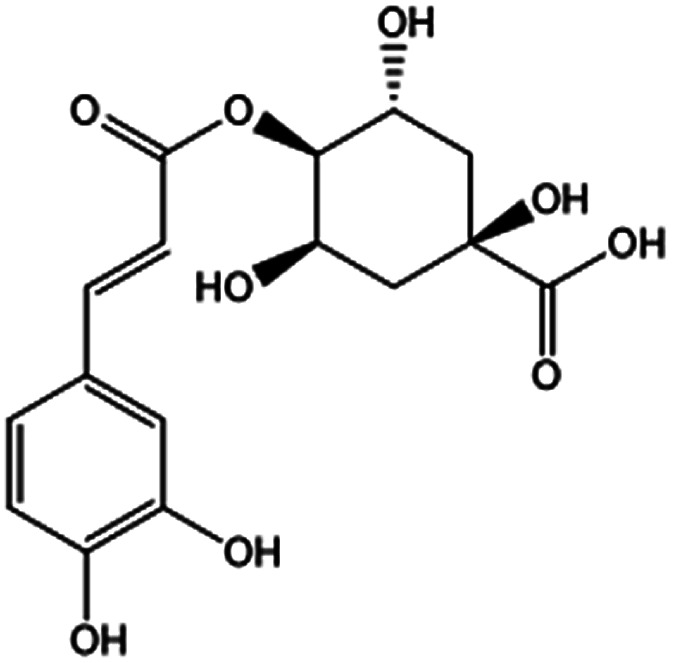
|
| KDZI-3 | 5-Caffeoylquinic acid | C16H18O9 | 354.31 |
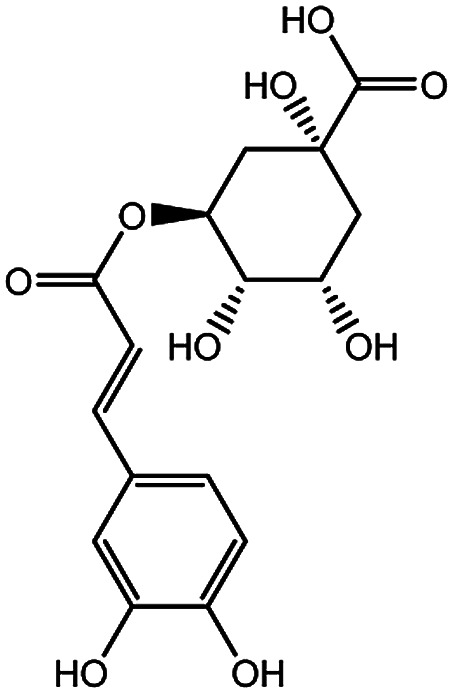
|
| KDZI-4 | Apigenin-7-O-β-d-glucuronopyranoside | C21H20O11 | 448.38 |
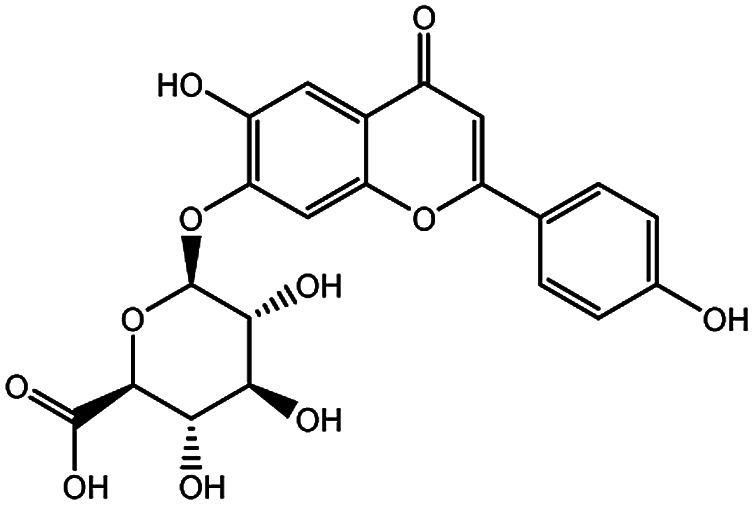
|
| KDZI-5 | Caffeic acid | C9H8O4 | 180.16 |
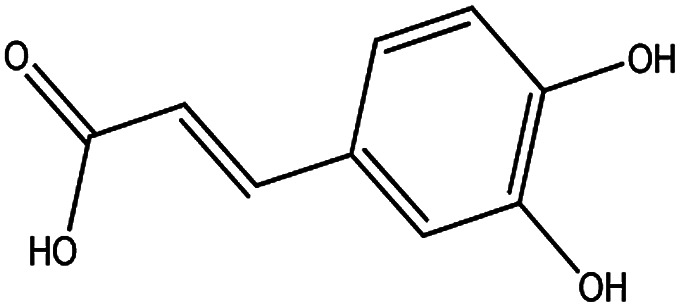
|
| KDZI-6 | Chicoric acid | C22H18O12 | 474.37 |
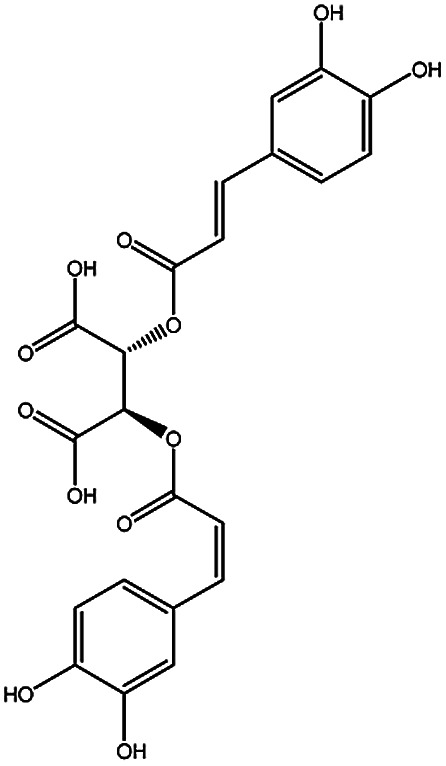
|
| KDZI-7 | Caftaric acid | C13H12O9 | 312.23 |
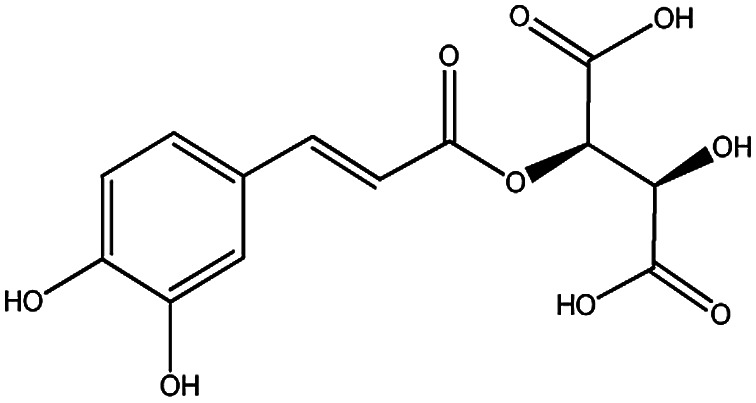
|
| KDZI-8 | Luteolin-7-O-β-d-gentiobioside | C27H30O16 | 610.52 |
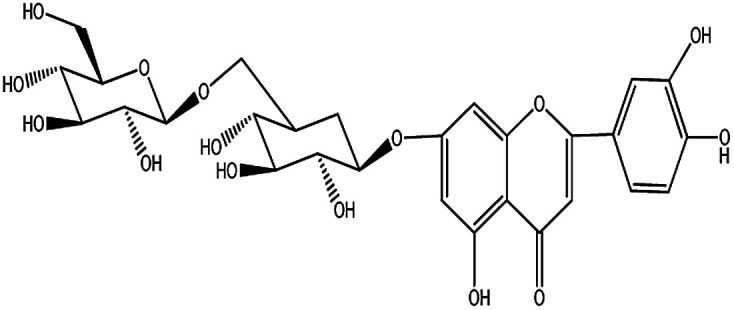
|
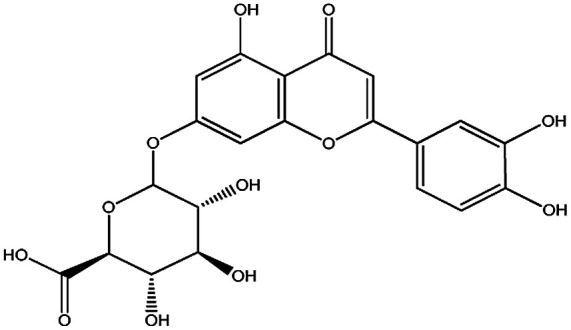
| KDZI-9 | Luteolin-7-O-β-d-glucopyranoside | C21H20O11 | 448.38 |
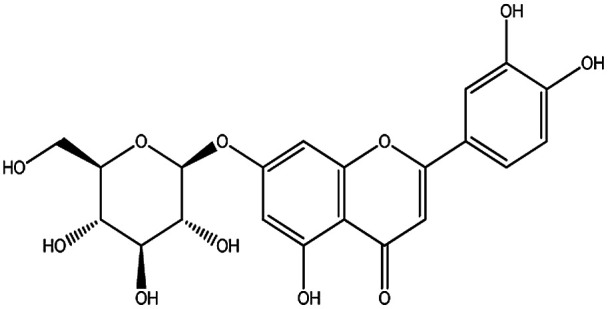
|
| KDZI-10 | Luteolin-7-O-β-d-glucuronide | C21H18O12 | 462.36 |
|
3.2. Targets fishing for KDZI
In the present study, 39 targets related to CVDs were predicted using the online server PharmMapper and their detailed information is described in ESI Table S1.†
3.3. Molecular docking
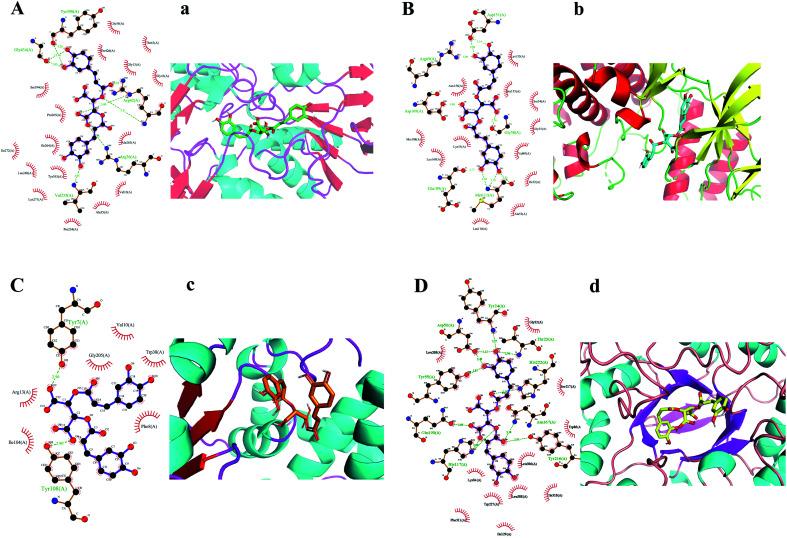
KDZI-6 was found possessing the largest docking scores for numerous target proteins among the ten ingredients. Thus, taking KDZI-6 as a typical objective, the docking results were introduced next. The three-dimensional structure and the schematic 2D dimensional of KDZI-6 with its best docking state in the active site of the proteins MAOB (PDB ID: 1OJ9), MAPK8 (PDB ID: 4UX9), GSTP1 (PDB ID: 1KBN) and AKR1C1 (PDB ID: 6IJX) are displayed in Fig. 2, and the binding energies are −11.2, −9.3, −6.7 and −10.6 kcal mol−1, respectively. In particular, a lower binding energy predicts a strong capacity between the ingredients and the target proteins. In addition, the comparison of the results of above-mentioned methods for the top 10 docking also exhibited in Table S2.† For the docking of MAOB, Fig. 2(A) and (a) showed the KDZI-6 stabilized by the nine hydrogen bonds involving the amino acid residues Tyr398, Gly434, Arg42, Arg36 and Val235 in MAOB, respectively. Also, it also interacted with 16 amino acid residues of MAOB via hydrophobic forces. Similarly, from Fig. 2(B) and (b), it can be seen that KDZI-6 is embedded to the active site of MAPK8. Nine hydrogen bonds were established between the ingredient and the protein referring to the six amino acid residues, Asp151, Arg69, Asp169, Gly38, Glu109 and Met111. It was also adjacent to twelve amino acid residues via hydrophobic interactions. Moreover, KDZI-6 possessed a good fit for the active sites of GSTP1, as depicted in Fig. 2(C) and (c). Two hydrogen bonds were found in the docking pose referring to the amino acid residues Tyr7 and Tyr108 in GSTP1. At the same time, there are hydrophobic interactions between the ligand and the amino acid residues Val10, Gly205, Trp38, Arg13, Ile104 or Phe8. The representation of the binding interaction of KDZI-6 with the active site of AKR1C1 is exhibited in Fig. 2(D) and (d). KDZI-6 was bound to the amino acid residues Tyr24, Asp50, Thr23, His222, Tyr55, Gln190, Asn167, His117 and Tyr216 though eleven hydrogen bonds, and the hydrophobic interactions involving eleven others amino acid residues in AKR1C1. The docking results not only showed the active sites in target proteins and the interactive force, but suggested that KDZI-6 might be of great significance for the development of new drugs as a multi-target ingredient in the future.
Fig. 2. Hydrogen bonds and hydrophobic interactions (A–D). Best-docked conformations (a–d).
3.4. Annotation analysis for the biological function of potential targets
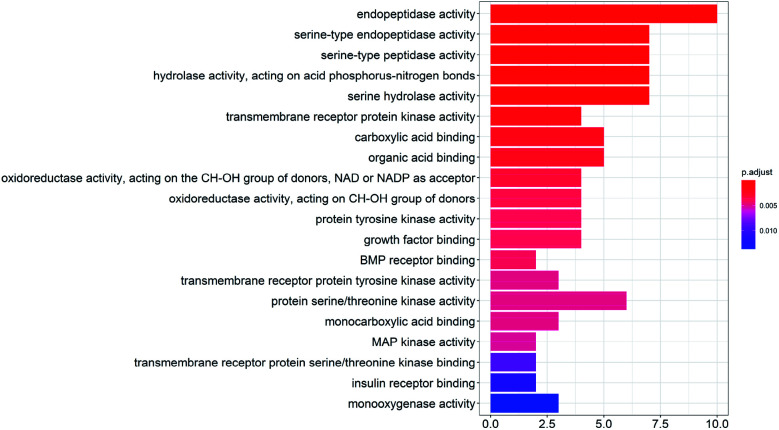
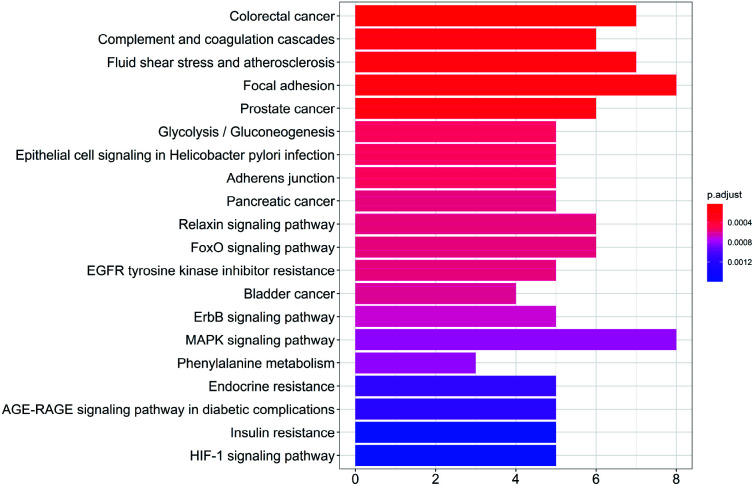
To study the biological functions of the potential targets, the GO enrichment analysis and KEGG analysis were performed in this research. Fig. 3 and 4 show the top 20 crucially significant GO molecular function annotations and pathways (p value < 0.01), respectively. The results of GO enrichment mainly included endopeptidase activity, serine-type endopeptidase activity, serine-type peptidase activity, hydrolase activity, acting on acid phosphorus–nitrogen bonds, serine hydrolase activity, transmembrane receptor protein kinase activity, carboxylic acid binding and organic acid binding. As shown in Fig. 4, the KEGG pathway analysis shows that the potential targets are intensively associated to the pathways as follows: colorectal cancer, complement and coagulation cascades, fluid shear stress and atherosclerosis, focal adhesion, prostate cancer, glycolysis/gluconeogenesis, epithelial cell signaling in Helicobacter pylori infection, adherens junction, pancreatic cancer.
Fig. 3. GO enrichment entries in the top 20 (p < 0.01).
Fig. 4. KEGG pathway enrichment entries in the top 20 (p < 0.01).
3.5. PPI and compound-target-pathway network construction
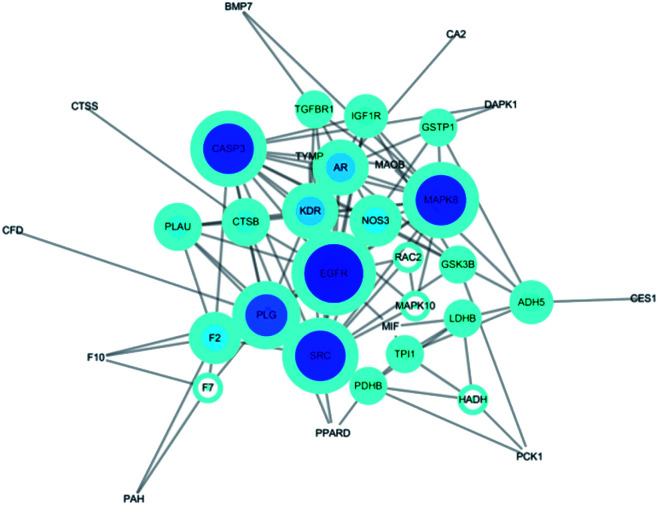
The potential protein interaction relationship among the targets was constructed (Fig. 5). There were 36 nodes and 202 edges after hiding disconnected nodes. Among them the targets CASP3 (degree: 14; betweenness: 0.14713729), PLG (degree: 12; betweenness: 0.13976426), MAPK8 (degree: 14; betweenness: 0.1048131), EGFR (degree: 16; betweenness: 0.20036317), SRC (degree: 14; betweenness: 0.12330768) were probably the most important roles in this protein interaction networks.
Fig. 5. Protein–Protein-Interaction Network. The node size in the network correlates with the degree of connectivity.
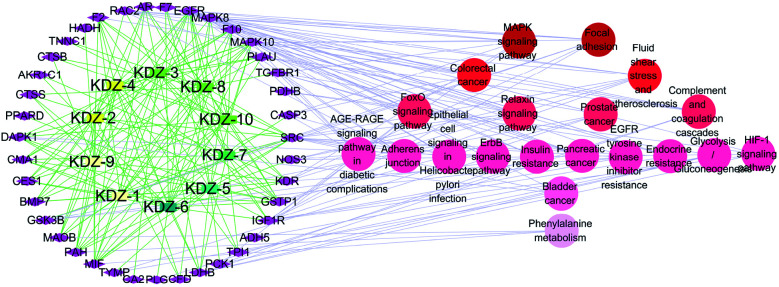
The Compound-Target-Pathway network was constructed by the connection of potential pathways and corresponding targets (Fig. 6), which resulted in 69 nodes (10 compounds, 39 targets, 20 signal pathways) and 554 edges. The possible interactions between compounds and target proteins were also evaluated with connection degree and the betweenness. The targets EGFR (degree: 24; betweenness: 0.15889369), MAPK10 (degree: 20; betweenness: 0.09662042), SRC (degree: 19; betweenness: 0.07250202), MAPK8 (degree: 15; betweenness: 0.0466211), GSTP1 (degree: 12; betweenness: 0.02678676), IGF1R (degree: 12; betweenness: 0.03521009) and MIF (degree: 11; betweenness: 0.02760243) have higher connection degrees and betweenness in the network. Moreover, the interactions between the identified compounds (KDZI-1 to KDZI-10) and the key targets (EGFR, MAPK10, SRC, MAPK8, GSTP1, IGF1R and MIF) were also modeled by both docking softwares, namely Sybyl-X and Autodock Vina. The docking scores are exhibited in Table 3. Comparatively, the bonds between KDZI-6 and EGFR, KDZI-8 and MAPK10, KDZI-6 and SRC, KDZI-6 and MAPK8, KDZI-6 and GSTP1, KDZI-6 and IGF1R, and KDZI-6 and MIF have better performance with higher the total score and lower binding energy. Most of these hub target proteins interacted with the KDZI-6 compound well, which also provided evidence to support that KDZI-6 might be a potential multi-target active compound and deserves to be further investigated and developed as a drug candidate.
Fig. 6. Compound-Target-Pathway (C-T-P) network. Rectangle nodes represented compounds, which go better in a clock wise according to color; diamond nodes represented targets; circle nodes represented pathways.
Docking scores of the hub targets with the main active ingredients of KDZI.
| Sybyl-X/Vina (kcal mol−1) | KDZI-1 | KDZI-2 | KDZI-3 | KDZI-4 | KDZI-5 | KDZI-6 | KDZI-7 | KDZI-8 | KDZI-9 | KDZI-10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EGFR | Total score | 7.7 | 5.65 | 5.59 | 5.6 | 5.37 | 8.12 | 5.18 | 7.86 | 7.36 | 7.86 |
| Binding energy | −7.5 | −7.1 | −7.4 | −8.1 | −6 | −7.8 | −6.8 | −8.4 | −8.6 | −8.4 | |
| MAPK10 | Total score | 6.8 | 6.11 | 5.4 | 5.8 | 4.18 | 8.29 | 5.53 | 9.19 | 7.78 | 5.68 |
| Binding energy | −8.4 | −8.4 | −8.4 | −9.7 | −6.3 | −9 | −7.4 | −9.8 | −9.7 | −9.9 | |
| SRC | Total score | 7.36 | 6.44 | 7.58 | 6.31 | 5.51 | 9.8 | 9.95 | 1.81 | 9.12 | 8.8 |
| Binding energy | −8.2 | −7.9 | −8.9 | −9.5 | −6.7 | −8.7 | −7.5 | −9.8 | −10.1 | −9.7 | |
| MAPK8 | Total score | 6.89 | 5.93 | 5.72 | 5.47 | 5.35 | 10.32 | 5.09 | 7.09 | 8.64 | 7.03 |
| Binding energy | −8.7 | −8.7 | −9 | −9.1 | −6.5 | −9.3 | −7.8 | −10.5 | −10 | −9.3 | |
| GSTP1 | Total score | 3.81 | 2.92 | 5.36 | 6.16 | 5.58 | 9.53 | 6.63 | 4.55 | 7.42 | 6.18 |
| Binding energy | −6.5 | −6.4 | −6.8 | −8 | −5.7 | −6.7 | −5.8 | −7.4 | −7.6 | −7.1 | |
| IGF1R | Total score | 4.25 | 4.04 | 4.22 | 5.08 | 4.66 | 8.7 | 3.8 | 5.61 | 4.97 | 3.8 |
| Binding energy | −7.2 | −6.9 | −7.2 | −7.6 | −5.8 | −8 | −6.3 | −8 | −8.3 | −8.2 | |
| MIF | Total score | 6.47 | 4.22 | 8.01 | 1.17 | 6.01 | 11.39 | 8.93 | 3.05 | 3.69 | 0.6 |
| Binding energy | −5.8 | −5.8 | −6 | −6.5 | −5.5 | −5.8 | −5.3 | −5.8 | −6.7 | −6.7 |
Meanwhile, as shown in Fig. 5, the target EGFR and MAPK pathways have the highest network connection degree, which could suggest the significance of the EGFR protein and MAPK pathway for the KDZI to exert its anti-CVD therapeutic effects. Studies have shown that myocardial injuries caused by ischemia is associated with hostile factors such as reactive oxygen species (ROS), inadequate angiogenesis and inflammation.32 ROS as one of the largest hostile factors generated in ischemic surroundings could directly injure cardiomyocytes and vascular cells and further impair myocardial function.33–36 High concentrations of ROS promote cell apoptosis by inducing the activation of JNKs and members of the MAPK family.37 Alternatively, the inhibition of JNKs would attenuate the myocardial ischemic apoptosis.38
The epidermal growth factor (EGF) is also known to activate Src and MAPK pathways.39 Therefore, the down-regulation of the epidermal growth factor receptor (EGFR) and MAPK family might be beneficial in preventing and treating myocardial ischemia. According to these results, it can be deduced that the active ingredients of KDZI could exert their anti-CVD effects via binding to the EGFR and inhibiting the MAPK pathway.
Cellular apoptosis is another critical event mediating the impairment of cardiovascular function.40–42 In the focal adhesion pathway, the expression of the adhesion molecule is a prerequisite for neutrophil-endothelium adherence, which in turn contributes to myocardial ischemia-reperfusion injuries.43 Focal adhesion kinase (FAK), a key enzyme in the integrin-mediated signal transduction process of adhesion molecules, promotes cell survival. Src kinase plays an upstream role in mediating the activation of known adhesion, survival, and stress-activated signaling pathways in response to oxidative stress. Both FAK and Src are of great significance in mediating the cell-matrix adhesion.44 In the present study, the target Src and the focal adhesion pathway also showed high connection degree, suggesting their considerable roles on anti-CVDs for the intervention of the multiple active ingredients of KDZI.
In addition, MIF elevation could respond to myocardial ischemia, which might be a useful early and specific diagnostic biomarker for myocardial ischemia.45 GSTP decreases the sensitivity of the heart caused by ischemic injury and IGF-1R takes precautions against the detrimental effects of myocardial infarction. Both of them have a protective effect on the myocardium.46–48 In general, KDZI might correlate with these targets, as shown in Fig. 6 and their related pathways in the treatment of CVDs. Further experiments including western blot, enzyme-linked immunosorbent, and polymerase chain reaction assays should be required to verify the present findings from a systematic aspect in the future.
4. Conclusions
As a traditional Chinese medicine injection, KDZI was extensively applied in clinical medicine for invigorating blood circulation for many years. Virtual screening and an integrated systems pharmacology approach in this study was used to evaluate the relationship between identified compounds and predicted targets against CVDs. GO molecular function and KEGG pathway enrichment analysis, constructions of Compound-Target-Pathway network and molecular docking were further performed. A total of 39 CVD-related targets of KDZI were predicted. The targets of EGFR, MAPK10, SRC, MAPK8, GSTP1, IGF1R and MIF and the signal pathways MAPK, focal adhesion, complement and coagulation cascades, fluid shear stress and atherosclerosis, prostate cancer, glycolysis/gluconeogenesis, epithelial cell signaling in Helicobacter pylori infection, adhesion junction, and pancreatic cancer could involve in the exerting of anti-CVD therapeutic effect of KDZI. In conclusion, this research provides a scientific basis to clarify the comprehensive pharmacological mechanism of KDZI acting on CVDs, and it could provide valuable clues on considering the active ingredient KDZI-6 as a multi-targets leading compound.
Conflicts of interest
There is no conflict of interest of any authors in relation to the submission.
Supplementary Material
Acknowledgments
This work was supported by National Nature Science Foundation of China (81973558), the Fujian Provincial Natural Science Foundation (2018J01596), Joint Funds for the Innovation of Science and Technology, Fujian province (2017Y9123 and 2019Y9068), Program for New Century Excellent Talents in Fujian Province University (2018), and Training program for excellent scientific research talents of young teachers in Fujian Province University (2017).
Electronic supplementary information (ESI) available: Two tables that reports potential targets information of KDZI and comparation of molecular docking results in the top 10 were listed. See DOI: 10.1039/d0ra07038f
References
- Zhou H. He L. Xu G. S. Chen L. X. Clin. Chim. Acta. 2020;507:210–218. doi: 10.1016/j.cca.2020.04.033. [DOI] [PubMed] [Google Scholar]
- Arnett D. K. Blumenthal R. S. Albert M. A. et al. . Circulation. 2019;140:E596–E646. doi: 10.1161/CIR.0000000000000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. C. Collins A. Ferrari R. et al. . J. Am. Coll. Cardiol. 2012;60:2343–2348. doi: 10.1016/j.jacc.2012.08.962. [DOI] [PubMed] [Google Scholar]
- Deng J. L. Guo M. Y. Li G. P. Xiao J. J. Gene Ther. 2020;27:360–369. doi: 10.1038/s41434-020-0148-6. [DOI] [PubMed] [Google Scholar]
- Amin S. Khan H. Curr. Bioact. Compd. 2016;12:103–106. doi: 10.2174/1573407212666160314195845. [DOI] [Google Scholar]
- Amin S. Ullah B. Ali M. Rauf A. Khan H. Uriarte E. Sobarzo-Sanchez E. Molecules. 2019;24:427. doi: 10.3390/molecules24030427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H. Amin S. Patel S. Life Sci. 2018;196:18–27. doi: 10.1016/j.lfs.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Khan H. Nabavi S. M. Sureda A. Mehterov N. Gulei D. Berindan-Neagoe I. Taniguchi H. Atanasov A. G. Eur. J. Med. Chem. 2018;153:29–33. doi: 10.1016/j.ejmech.2017.10.065. [DOI] [PubMed] [Google Scholar]
- Khan H. Sureda A. Belwal T. Cetinkaya S. Suntar I. Tejada S. Devkota H. P. Ullah H. Aschner M. Autoimmun. Rev. 2019;18:647–657. doi: 10.1016/j.autrev.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi-Rad M. Ozcelik B. Altin G. et al. . Trends Food Sci. Technol. 2018;80:242–263. doi: 10.1016/j.tifs.2018.08.008. [DOI] [Google Scholar]
- Mishra A. P. Saklani S. Salehi B. Parcha V. Sharifi-Rad M. Milella L. Iriti M. Sharifi-Rad J. Srivastava M. Cell. Mol. Biol. 2018;64:35–43. doi: 10.14715/cmb/2018.64.8.6. [DOI] [PubMed] [Google Scholar]
- Liu X. M. Tao Y. Wang F. L. Yao T. Fu C. Zheng H. Yan Y. Liang X. Jiang X. N. Zhang Y. L. BMC Complementary Altern. Med. 2017;17:10. doi: 10.1186/s12906-016-1537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L. Zhang Z. Z. Hou Z. G. Yang B. Li A. Z. Guo X. J. Wang Y. M. Li Y. B. Anal. Methods. 2015;7:5210–5217. doi: 10.1039/C4AY03103B. [DOI] [Google Scholar]
- Zhang W. J. Huai Y. Miao Z. P. Qian A. R. Wang Y. H. Front. Pharmacol. 2019;10:23. doi: 10.3389/fphar.2019.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. Zhang X. W. Mao B. Jiang H. L. J. Ethnopharmacol. 2020;249:10. doi: 10.1016/j.jep.2019.112425. [DOI] [PubMed] [Google Scholar]
- Yao H. Huang X. M. Xie Y. J. Huang X. L. Ruan Y. J. Lin X. H. Huang L. Y. Shi P. Y. Front. Pharmacol. 2018;9:16. doi: 10.3389/fphar.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q. Q. Liu Y. Liu C. H. Liu L. Meng D. L. Bioorg. Chem. 2019;91:15. doi: 10.1016/j.bioorg.2019.103118. [DOI] [PubMed] [Google Scholar]
- Wang Y. Sun Y. W. Wang Y. M. Ju Y. Meng D. L. Bioorg. Chem. 2019;88:9. doi: 10.1016/j.bioorg.2019.102924. [DOI] [PubMed] [Google Scholar]
- Yu G. H. Luo Z. Q. Zhou Y. T. Zhang L. Wu Y. Ding L. Shi Y. Y. Biomed. Pharmacother. 2019;117:11. doi: 10.1016/j.biopha.2019.109094. [DOI] [PubMed] [Google Scholar]
- Liu X. F. Ouyang S. S. Yu B. Liu Y. Huang K. Gong J. Y. Zheng S. Y. Li Z. H. Jiang H. L. Nucleic Acids Res. 2010;38:W609–W614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Pan C. X. Gong J. Y. Liu X. F. Li H. L. J. Chem. Inf. Model. 2016;56:1175–1183. doi: 10.1021/acs.jcim.5b00690. [DOI] [PubMed] [Google Scholar]
- Wang X. Shen Y. H. Wang S. W. Li S. L. Zhang W. L. Liu X. F. Lai L. H. Pei J. F. Li H. L. Nucleic Acids Res. 2017;45:W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. K. Hu Y. C. Wang L. C. Wang Y. W. Na S. S. Wang J. Shun Y. H. Wang X. Q. Xue P. F. Zhao P. W. Su L. P. J. Evidence-Based Complementary Altern. Med. 2018;2018:7956503. doi: 10.1155/2018/7956503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Lai L. T. Ju Y. Liu C. H. Meng D. L. Bioorg. Chem. 2019;92:15. doi: 10.1016/j.bioorg.2019.103302. [DOI] [PubMed] [Google Scholar]
- SYBYL Molecular modeling software, Version 1.3, Tripos Inc., St. Louis, MO, USA, 2007 [Google Scholar]
- Trott O. Olson A. J. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsek K. Mavri J. Dolenc M. S. Gobec S. Turk S. J. Chem. Inf. Model. 2014;54:1254–1267. doi: 10.1021/ci400649p. [DOI] [PubMed] [Google Scholar]
- Schrodinger L. L. C., The PyMOL Molecular Graphics System, Version 1.8, 2015
- Laskowski R. A. Swindells M. B. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Yu G. Wang L. G. Han Y. He Q. Y. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M. E. Ono K. Ruscheinski J. Wang P. L. Ideker T. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. Cha M. J. Song B. W. Kim I. K. Chang W. Lim S. Choi E. J. Ham O. Lee S. Y. Chung N. Jang Y. Hwang K. C. Stem Cell. 2010;28:555–563. doi: 10.1002/stem.302. [DOI] [PubMed] [Google Scholar]
- Dhalla N. S. Elmoselhi A. B. Hata T. Makino N. Cardiovasc. Res. 2000;47:446–456. doi: 10.1016/S0008-6363(00)00078-X. [DOI] [PubMed] [Google Scholar]
- Fan H. Sun B. Gu Q. Lafond-Walker A. Cao S. Becker L. C. Am. J. Physiol. Heart Circ. Physiol. 2002;282:H1778–H1786. doi: 10.1152/ajpheart.00796.2000. [DOI] [PubMed] [Google Scholar]
- Kloner R. A. Jennings R. B. Circulation. 2001;104:3158–3167. doi: 10.1161/hc5001.100039. [DOI] [PubMed] [Google Scholar]
- Ren G. Dewald O. Frangogiannis N. G. Curr. Drug Targets - Inflamm. Allergy. 2003;2:242–256. doi: 10.2174/1568010033484098. [DOI] [PubMed] [Google Scholar]
- Lee H. S. Hwang C. Y. Shin S. Y. Kwon K. S. Cho K. H. Sci. Signal. 2014;7:ra52. doi: 10.1126/scisignal.2005260. [DOI] [PubMed] [Google Scholar]
- Inagaki K. Hahn H. S. Dorn G. W. Mochly-Rosen D. Circulation. 2003;108:869–875. doi: 10.1161/01.CIR.0000081943.93653.73. [DOI] [PubMed] [Google Scholar]
- Clemente L. Boeldt D. S. Grummer M. A. Morita M. Morgan T. K. Wiepz G. J. Bertics P. J. Bird I. M. Mol. Cell. Endocrinol. 2020;499:14. doi: 10.1016/j.mce.2019.110590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akki A. Zhang M. Murdoch C. Brewer A. Shah A. M. J. Mol. Cell. Cardiol. 2009;47:15–22. doi: 10.1016/j.yjmcc.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Brown D. I. Griendling K. K. Free Radic. Biol. Med. 2009;47:1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerries C. Grote K. Hilfiker-Kleiner D. Circ. Res. 2007;100:894–903. doi: 10.1161/01.RES.0000261657.76299.ff. [DOI] [PubMed] [Google Scholar]
- Vinten-Johansen J. Cardiovasc. Res. 2004;61:481–497. doi: 10.1016/j.cardiores.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Hui A. Y. Meens J. A. Schick C. Organ S. L. Qiao H. Tremblay E. A. Schaeffer E. Uniyal S. Chan B. M. C. Elliott B. E. J. Cell. Biochem. 2009;107:1168–1181. doi: 10.1002/jcb.22219. [DOI] [PubMed] [Google Scholar]
- Fan F. Dart A. M. J. Am. Coll. Cardiol. 2015;66:C131–C132. doi: 10.1016/j.jacc.2015.06.507. [DOI] [Google Scholar]
- Conklin D. J. Guo Y. R. Jagatheesan G. Kilfoil P. J. Haberzettl P. Hill B. G. Baba S. P. Guo L. P. Wetzelberger K. Obal D. Rokosh D. G. Prough R. A. Prabhu S. D. Velayutham M. Zweier J. L. Hoetker J. D. Riggs D. W. Srivastava S. Bolli R. Bhatnagar A. Circ. Res. 2015;117:437–449. doi: 10.1161/CIRCRESAHA.114.305518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki C. Otani H. Sato D. Okada T. Hattori R. Imamura H. Int. J. Cardiol. 2010;138:9–18. doi: 10.1016/j.ijcard.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Huynh K. McMullen J. R. Julius T. L. Tan J. W. Love J. E. Cemerlang N. Kiriazis H. Du X. J. Ritchie R. H. Diabetes. 2010;59:1512–1520. doi: 10.2337/db09-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.