Comparison of computed relative MM interaction energies of known inhibitors to the SARS-CoV-2 Mpro deriving from crystal structures 6Y2F and 6LU7 (ref. 12 and 14).
| Inhibitor | Formula: P5–P4–P3–P2–P1–P1′ | ΔΔEint,MMa [kcal mol−1] | M w b [g mol−1] | IC50expc SARS-CoV (2003) Mpro [μM] | IC50expd SARS-CoV-2 (2019/20) Mpro [μM] |
|---|---|---|---|---|---|
| 13ae,f |
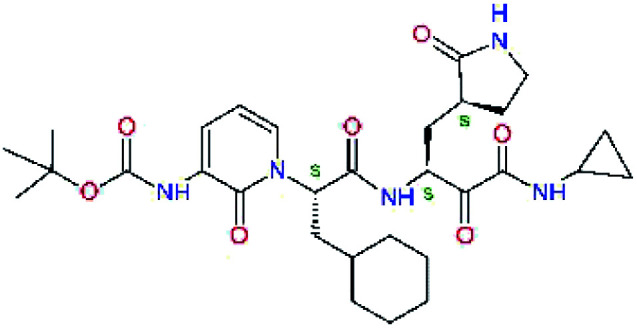
|
4.4 | 583.7 | — | 2.39 |
| 13bf |
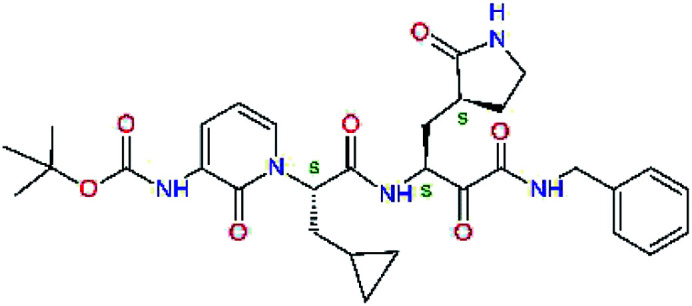
|
0.0i | 591.7 | 0.90 | 0.67 |
| N3g |
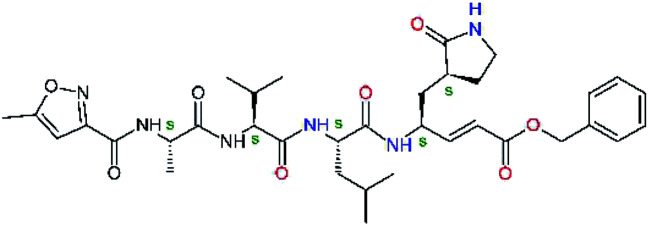
|
−4.1 | 680.8 | — | — |
| 11nh |
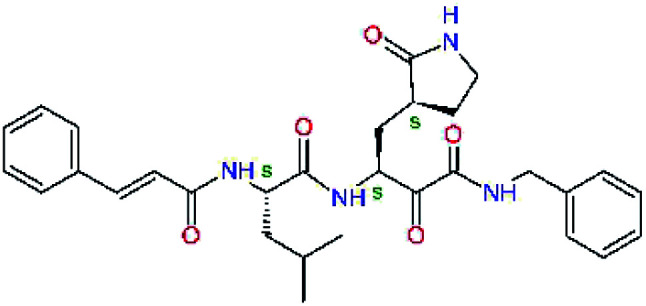
|
7.6 | 532.6 | 0.33 | — |
| 11rf,h |
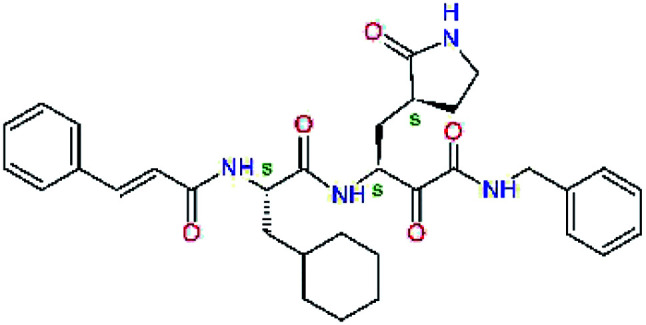
|
5.6 | 572.7 | 0.71 | 0.18 |
Relative interaction energy taken with respect to the reference inhibitor 13b was calculated by molecular mechanics (MM-OPLS3e) in solution: ΔΔEint,MM = ΔEint,MM(Ix) − ΔEint,MM(13b) = [Etot,MM{Mpro–Ix}aq − Etot,MM{Mpro}aq − Etot,MM{Ix}aq] − ΔEint,MM(13b), where Etot,MM is total energy of solvated enzyme-inhibitor complex {Mpro–Ix}aq, solvated enzyme {Mpro}aq, or solvated inhibitor {Ix}aq.35–38 The relative interaction energy ΔΔEint,MM describes changes in bonding and non-bonding components of potential energy of the enzyme and inhibitor upon the enzyme-inhibitor complex formation.
Molar mass.
Half-maximal inhibitory concentration determined in enzyme-inhibition assay for the Mpro of SARS-CoV from the 2003 outbreak.14,23
Half-maximal inhibitory concentration determined in enzyme-inhibition assay for the Mpro of SARS-CoV-2 from the 2019/20 outbreak.12
Interaction energy of irreversible Michael acceptor or α-ketoamide inhibitors was computed after breaking the covalent bond of the P1 residue to the catalytic Cys145.
Taken from ref. 12.
Taken from ref. 14.
Taken from ref. 23.
The 13b was used as the reference inhibitor in all calculations of the relative interaction energy ΔΔEint,MM.
