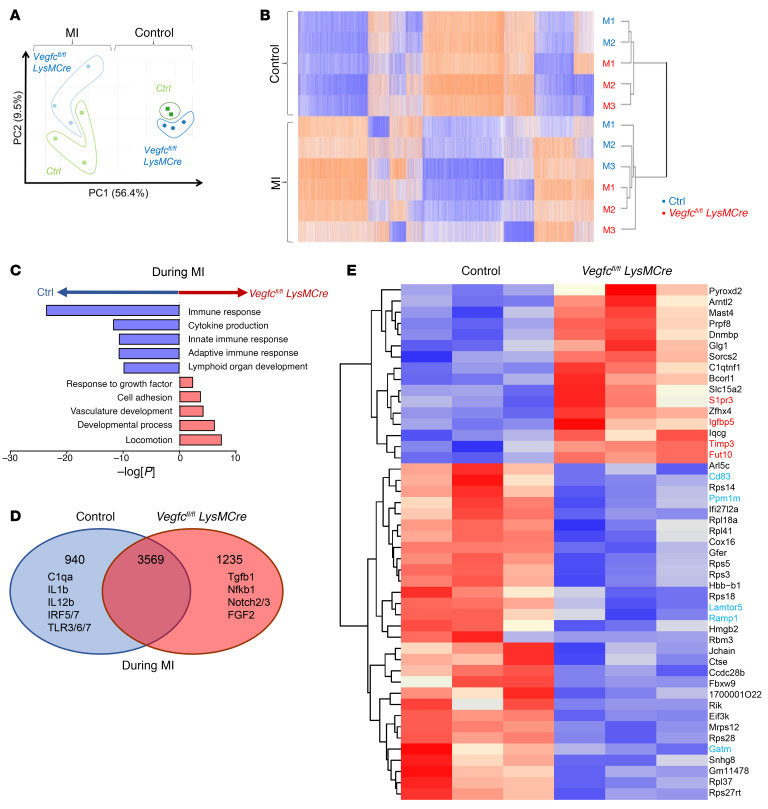
Figure 8. Impaired immune response with myeloid Vegfc deficiency after MI.
Experimental C57BL/6 or Vegfcfl/fl versus Vegfcfl/fl LysMCre mice were subjected to coronary artery occlusion, and bulk mRNA gene expression analysis was performed for the LV. (A) Principal component analysis (PCA) revealing MI as a main source of variance in the data set. Data for the nonligated animals were clustered together, consistent with relatively comparable gene expression profiles at steady state. (B) Heatmap analysis and hierarchical clustering revealed distinct changes between nonligated and ligated animals at the transcriptional level. M1, M2, and M3 represent individual animals used for each group. (C) Gene ontology pathway interrogation revealed significant downregulation of immune response genes in the absence of myeloid Vegfc. In contrast, developmental pathways were induced in Vegfcfl/fl LysMCre mice, consistent with a hypertrophic response. (D) Venn diagram of differentially expressed or shared expression genes. (E) Heatmap of normalized top 50 absolute log fold changes in Vegfcfl/fl LysMCre mice compared with controls after MI. Genes highlighted in red are associated with inflammation and fibrosis, whereas those in cyan are associated with a lymphatic response and inflammation resolution.