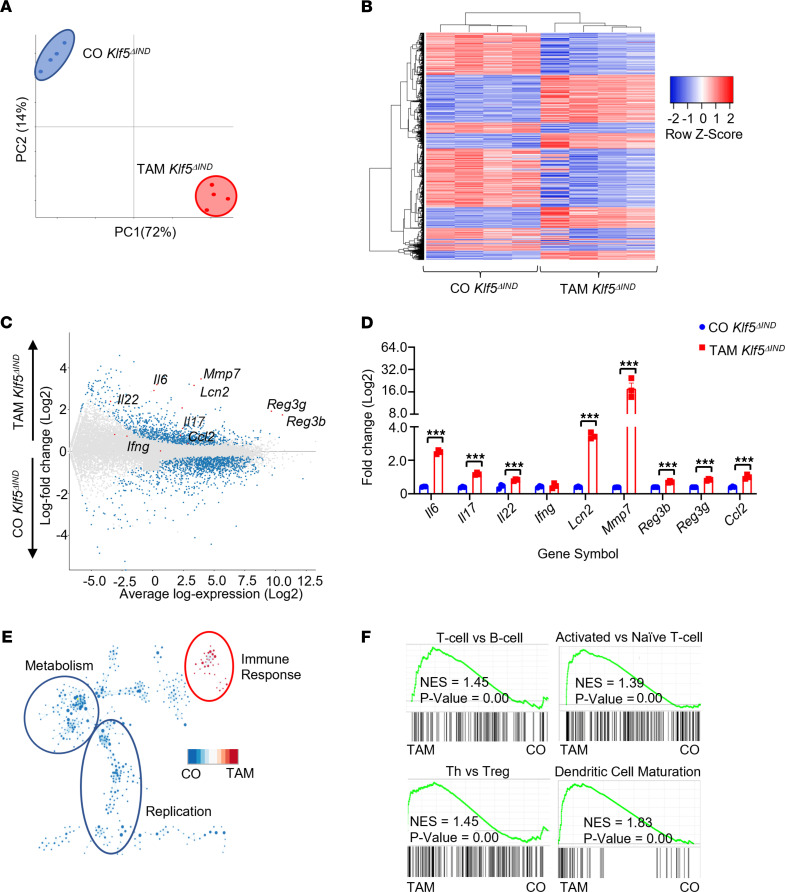
Figure 3. Deletion of Klf5 from intestinal epithelial cells results in changes to the global transcriptomic pathways to inflammatory responses.
(A) Principal component analysis of bulk RNA-Seq data. Comparison between CO- and TAM-treated Klf5ΔIND mice (n = 4). PC1 = 72% and PC2 = 14%. (B) Heatmap representation of significant differentially expressed genes from DESeq2. (C) MA-plot analysis of all genes from bulk RNA-Seq. Blue points represent a significant differentially expressed gene with a cutoff of P = 0.05. Red points are representative genes of interest with known roles in intestinal inflammation and regeneration. (D) RT-qPCR confirming genes of interest from bulk RNA-Seq analysis. (E) Leading-edge analysis after GSEA pathway analysis. Blue represents pathways correlated to CO-treated Klf5ΔIND mice and red represents pathways correlated with TAM-treated Klf5ΔIND mice. (F) GSEA pathway analysis positively correlated with TAM-treated Klf5ΔIND mice. Data from graphs represent mean ± SEM. ***P < 0.001; unpaired 1-tailed t test.