Abstract
Introduction
Computed tomography is useful for the diagnosis of coronavirus disease (COVID‐19) pneumonia. However, many types of interstitial lung diseases and even bacterial pneumonia can show abnormal chest shadows that are indistinguishable from those observed in COVID‐19 pneumonia. Thus, it is necessary to identify useful biomarkers that can efficiently distinguish COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases. Herein, we investigated the usefulness of serum Krebs von den Lungen 6 (KL‐6) and surfactant protein D (SP‐D) for identifying patients with COVID‐19 pneumonia among patients with abnormal chest shadows consistent with COVID‐19 pneumonia.
Method
This was a retrospective cohort study of consecutive patients who underwent evaluation of serum KL‐6 and SP‐D at a single center from February 2019 to December 2020. A total of 54 patients with COVID‐19 pneumonia and 65 patients with COVID‐19 pneumonia‐like diseases were enrolled in this study from the source population. Serum KL‐6 and SP‐D levels in both groups were analyzed.
Result
The serum levels of KL‐6 and SP‐D in patients with COVID‐19 pneumonia were significantly lower than those in patients with COVID‐19 pneumonia‐like disease (median [interquartile range]: 208.5 [157.5–368.5] U/ml vs. 430 [284.5–768.5] U/ml, p < 0.0001 and 24.7 [8.6–51.0] ng/ml vs. 141 [63.7–243.5] ng/ml, p < 0.0001, respectively). According to receiver operating characteristic (ROC) analysis, the areas under the ROC curves (95% confidence intervals) of serum KL‐6 and SP‐D levels for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases were 0.761 (0.675–0.847) and 0.874 (0.812–0.936), respectively. The area under the ROC curve of serum SP‐D was significantly larger than that of serum KL‐6 (p = 0.0213), suggesting that serum SP‐D can more efficiently distinguish COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases.
Conclusion
Serum SP‐D is a promising biomarker for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases. Serum SP‐D can be useful for the management of patients with abnormal chest shadow mimicking COVID‐19 pneumonia.
Keywords: COVID‐19, diagnosis, KL‐6, SP‐D
1. INTRODUCTION
Coronavirus disease (COVID‐19) is a major health challenge worldwide. Several clinically important variants with greater transmissibility have emerged. The highly contagious nature of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a serious problem in the management of patients with COVID‐19. Early diagnosis of COVID‐19 is established by a positive result of nucleic acid amplification test (NAAT) or antigen test for SARS‐CoV‐2. However, the accuracy of NAAT and antigen testing is not perfect. False‐negative results are particularly serious because they can promote virus transmission and cause nosocomial infections. False‐negative results of NAAT are reported to occur in approximately 30% of patients with COVID‐19. 1 Moreover, the accuracy of antigen tests is less than that of NAAT 2 Therefore, it is important to suspect COVID‐19 by clinical presentation, isolate suspected patients and repeat NAAT or antigen testing. However, there are no clinical presentations characteristic of COVID‐19. The typical symptoms of COVID‐19, such as cough, fever, and fatigue, are quite common in other acute respiratory diseases, and a substantial number of patients are asymptomatic. 3 Thus, it is not easy to suspect COVID‐19 solely based on clinical presentation.
Chest computed tomography (CT) is known to have high sensitivity in diagnosing COVID‐19 and is used as a useful tool for the diagnosis of COVID‐19 in clinical practice. 4 The typical findings of chest CT are bilateral ground‐glass opacities (GGOs) and/or consolidation, predominantly located peripherally and posteriorly in the lungs. 5 However, the specificity of chest CT for diagnosing COVID‐19 is low. 4 Many types of interstitial lung diseases (ILD) frequently show similar shadows on chest CT and chest CT findings of ILD are sometimes indistinguishable from those of COVID‐19 pneumonia. In addition, even bacterial pneumonia can show GGOs, and bilateral or multilobar distribution is not rare. 6 Thus, when patients with negative NAAT results and abnormal chest shadow mimicking COVID‐19 pneumonia are admitted to the hospital, the possibility of false‐negative NAAT results must be considered. Such cases are not rare, as patients presenting bilateral GGOs and/or consolidation on chest CT are often admitted to the hospital, regardless of the etiology. Therefore, a biomarker that can distinguish COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases is required for infection control in COVID‐19 outbreak settings.
Serum surfactant protein D (SP‐D) and Krebs von den Lungen 6 (KL‐6) are useful biomarkers for the diagnosis and evaluation of ILD. 7 , 8 Both biomarkers are reported to increase in many types of ILD, such as idiopathic pulmonary fibrosis and collagen vascular disease‐related interstitial pneumonia (CVD‐IP). 7 In COVID‐19, serum KL‐6 and SP‐D levels were reported to increase in severe patients and were useful as prognostic biomarkers of severity. 9 , 10 , 11 , 12 , 13 However, their usefulness as diagnostic biomarkers remains unknown. Based on this knowledge, we addressed the usefulness of serum KL‐6 and SP‐D for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases by evaluating serum KL‐6 and SP‐D levels in COVID‐19 pneumonia patients and non‐COVID‐19 pneumonia patients whose chest CT images showed abnormal shadows mimicking COVID‐19 pneumonia on retrospective analysis.
2. MATERIALS AND METHODS
2.1. Study design and participants
Of all patients for whom serum KL‐6 and SP‐D levels were evaluated simultaneously in the Department of Respiratory Medicine, Tokyo Medical University Hospital from February 2019 to December 2020, patients with COVID‐19 pneumonia and patients with COVID‐19 pneumonia‐like diseases were enrolled in this study. Patients with COVID‐19 pneumonia were enrolled based on the following criteria: (1) patients with positive results of NAAT or antigen test for SARS‐CoV‐2; (2) patients presenting abnormal chest shadow on chest CT consistent with COVID‐19 pneumonia. Patients with COVID‐19 pneumonia‐like diseases were enrolled based on the following criteria: (1) patients in whom chest CT revealed bilateral GGOs and/or consolidation; (2) patients who took less than 30 days from clinical onset to the evaluation of serum KL‐6 and SP‐D; (3) patients with confirmed negative results of NAAT for SARS‐CoV‐2 or patients for whom the evaluation of serum KL‐6 and SP‐D was performed before January 16, 2020, which is when the first case of COVID‐19 was confirmed in Japan. Patients who met the following criteria were excluded: (1) those who were diagnosed with any ILD before the evaluation of serum KL‐6 and SP‐D and (2) those in whom chest CT revealed an abnormal shadow suggesting chronic fibrosing ILD, such as traction bronchiectasis and honeycomb change. A summary of patient enrollment is presented in Figure 1. Clinical measurements of patients with COVID‐19 pneumonia and COVID‐19 pneumonia‐like diseases were retrospectively analyzed.
Figure 1.
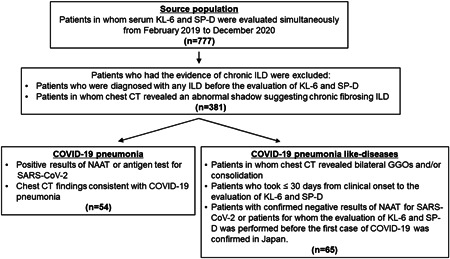
Flow chart of patient enrollment. CT, computed tomography; COVID‐19, coronavirus disease; GGOs, ground‐glass opacities; ILD, interstitial lung diseases; KL‐6, Krebs von den Lungen 6; NAAT, nucleic acid amplification test; SARS‐COV‐2, severe acute respiratory syndrome coronavirus 2; SP‐D, surfactant protein D
2.2. Data collection
Clinical data were obtained from patients’ medical records. Laboratory findings and chest CT findings at the time of the initial evaluation of serum KL‐6 and SP‐D levels were also recorded. Chest CT images were reviewed and interpreted by an experienced pulmonologist without knowledge of any relevant clinical information.
2.3. Measurements of serum KL‐6 and SP‐D
Serum levels of KL‐6 and SP‐D were measured using chemiluminescent enzyme immunoassay (CLEIA) with Lumipulse® G KL‐6 (Fujirebio Corporation) and CL SP‐D Yamasa NX (Yamasa Corporation), respectively. The coefficients of variation of Lumipulse® G KL‐6 and CL SP‐D Yamasa NX are <10% and <15%, respectively. Measurements were performed according to the manufacturer's instructions. The lower detection limits were set at 17.2 ng/ml and 50 U/ml for SP‐D and KL‐6, respectively.
2.4. Diagnosis
The diagnosis of COVID‐19 pneumonia was based on positive results of NAAT or antigen testing and chest CT findings consistent with COVID‐19 pneumonia. The diagnosis of drug‐induced pneumonia was based on a history of causative drug exposure and exclusion of other causes of lung injury. The diagnosis of bacterial pneumonia was based on positive results of bacterial sputum culture and/or clinical improvement with antibacterial agents. CVD‐IP was diagnosed based on the presence of underlying collagen vascular disease and exclusion of other causes of lung injury. Cryptogenic organizing pneumonia (COP) was diagnosed based on pathological findings consistent with organizing pneumonia and spontaneous or corticosteroid‐induced improvement and exclusion of other causes. The diagnosis of hypersensitivity pneumonitis (HP) was based on a history of causative antigen exposure and pathological findings consistent with HP and improvement with corticosteroids or physical isolation from the antigen. The diagnosis of acute interstitial pneumonia (AIP) was based on the rapidly progressive onset of clinical symptoms and exclusion of other causes of lung injury.
2.5. Statistical analyses
Variables are presented as medians and interquartile range (IQR). Statistical significance was set at p < 0.05. Categorical variables were compared using Fisher's exact test. Kruskal–Wallis test and Mann–Whitney U test were performed to analyze the differences between the selected groups. Serum SP‐D and KL‐6 levels below the lower detection limit were given half the limit's value. Multivariable logistic regression analysis and receiver operating characteristic (ROC) analyses were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan). The areas under the ROC curves were compared using the DeLong test. In multivariable logistic regression analysis, the diagnosis of COVID‐19 was used as a dependent variable, whereas age, sex, serum KL‐6 level, serum SP‐D level, and cancer were used as independent variables. All other statistical analyses were performed using GraphPad Prism software (GraphPad, San Diego, CA).
3. RESULTS
3.1. Study of patients' characteristics
We enrolled 54 patients with COVID‐19 pneumonia and 65 patients with COVID‐19 pneumonia‐like diseases. Two patients with COVID‐19 pneumonia initially had negative NAAT results and were diagnosed by repeating NAAT. There was no significant difference in the proportion of female patients in the group with COVID‐19 pneumonia and the group with COVID‐19 pneumonia‐like diseases (33% vs. 38%, p = 0.5726). Patients with COVID‐19 pneumonia were significantly younger than patients with COVID‐19 pneumonia‐like diseases (51 [38–64] years vs. 65 [53.5–73] years, p = 0.0009). The time between clinical onset and evaluation of SP‐D and KL‐6 was significantly shorter in patients with COVID‐19 pneumonia than in those with COVID‐19 pneumonia‐like diseases (7 [4–9] days vs. 10 [5–16] days, p = 0.036). Cancer prevalence was significantly higher among patients with COVID‐19 pneumonia‐like diseases than among those with COVID‐19 pneumonia (34% vs. 2%, p < 0.0001). There was no significant difference in the proportion of patients showing either GGOs or consolidation on chest CT between patients with COVID‐19 pneumonia and COVID‐19 pneumonia‐like diseases (85% vs. 91%, p = 0.4001 and 33% vs. 42%, p = 0.4481, respectively). There was also no significant difference in the number of affected lobes on chest CT between patients with COVID‐19 pneumonia and COVID‐19 pneumonia‐like diseases (5 [2.75–5] vs. 5 [4‐5], p = 0.1818). In blood tests, the white blood cell count of patients with COVID‐19 pneumonia was significantly lower than that of patients with COVID‐19 pneumonia‐like diseases (4850 [3675–6400]/μl vs. 7500 [6050–10,100]/μl, p < 0.0001). Among the patients with COVID‐19 pneumonia‐like diseases, 19 patients (29%) were diagnosed with drug‐induced pneumonia, 14 (22%) were diagnosed with bacterial pneumonia, eight (12%) with CVD‐IP, six (9%) with COP, five (8%) with HP, and five (8%) with AIP. Other patients were diagnosed with pneumocystis pneumonia (n = 3), diffuse alveolar hemorrhage (n = 2), radiation pneumonitis (n = 1), eosinophilic pneumonia (n = 1), and intravascular lymphoma (n = 1). The characteristics of the enrolled patients are summarized in Table 1.
Table 1.
Characteristics of patients enrolled in this study
| COVID‐19 pneumonia (n = 54) | COVID‐19 pneumonia‐like diseases (n = 65) | pValue | |
|---|---|---|---|
| Female (%) | 33 | 38 | 0.5726 |
| Age (years) | 51 (38–64) | 65 (53.5–73) | 0.0009 |
| Never smoker (%) | 46 | 46 | >0.9999 |
| Pack‐years | 0.9 (0–22) | 8.5 (0–25) | 0.4689 |
| Time between clinical onset and evaluation of SP‐D and KL‐6 (days) | 7 (4–9) | 10 (5–16) | 0.036 |
| Asymptomatic patients (%) | 6 | 6 | <0.9999 |
| Patients with respiratory failure (%) | 20 | 35 | 0.1024 |
| Patients with cancer (%) | 2 | 34 | <0.0001 |
| Chest CT findings | |||
| Ground‐glass attenuation(%) | 85 | 91 | 0.4001 |
| Consolidation (%) | 33 | 42 | 0.4481 |
| The number of affected lobes | 5 (2.75–5) | 5 (4–5) | 0.1818 |
| Blood test | |||
| WBC (μl) | 4850 (3675–6400) | 7500 (6050–10100) | <0.0001 |
| LDH (IU/L) | 243 (200–332) | 289 (204–421) | 0.1295 |
| BUN (mg/dl) | 13.2 (10.3–17.6) | 13.7 (10–18.4) | 0.8664 |
| Creatinine (mg/dl) | 0.74 (0.63–1.055) | 0.7 (0.56–0.92) | 0.0624 |
| CRP (mg/dl) | 2.6 (1.2–5.7) | 4.8 (1.7–9.9) | 0.0652 |
| Diagnosis (%) | |||
| Drug‐induced pneumonia | 29 | ||
| Bacterial pneumonia | 22 | ||
| CVD‐IP | 12 | ||
| COP | 9 | ||
| HP | 8 | ||
| AIP | 8 | ||
| Others | 12 |
Note: Data are presented as median (interquartile range). Significant results (p < 0.05) are shown in bold type.
Abbreviations: AIP, acute interstitial pneumonia; BUN, blood urea nitrogen; COP, cryptogenic organizing pneumonia; COVID‐19, coronavirus disease; CRP, c‐reactive protein; CT, computed tomography; CVD‐IP, collagen vascular disease‐related interstitial; pneumonia; HP, hypersensitivity pneumonitis; LDH, lactate dehydrogenase; WBC, white blood cell.
3.2. Serum KL‐6 and SP‐D levels in patients with COVID‐19 pneumonia and patients with COVID‐19 pneumonia‐like diseases
The levels of serum KL‐6 and SP‐D in patients with COVID‐19 pneumonia were significantly lower than those in patients with COVID‐19 pneumonia‐like disease (208.5 [157.5–368.5] U/ml vs. 430 [284.5–768.5] U/ml, p < 0.0001 and 24.7 [8.6–51.0] ng/ml vs. 141 [63.7‐243.5] ng/ml, p < 0.0001, respectively) (Figure 2). When analyzed after dividing patients based on the diagnosis, the serum SP‐D level of patients with COVD‐19 pneumonia was significantly lower than that of patients with drug‐induced pneumonia, bacterial pneumonia, COP, or AIP, while the serum KL‐6 level of COVID‐19 pneumonia was significantly lower than that of patients diagnosed with drug‐induced pneumonia or CVD‐IP. The median levels of serum SP‐D were 198 (106–356) ng/ml in drug‐induced pneumonia, 119 (51.5–222.5) ng/ml in bacterial pneumonia, 202.5 (86–309.3) ng/ml in COP, 63.7 (29.9–160.3) ng/ml in CVD‐IP, 78.7 (52.6–152.2) ng/ml in HP, 182 (64–1355) ng/ml in AIP, and 113 (44.8–325.8) ng/ml in others. The median levels of serum KL‐6 were 541 (299–904) U/ml in drug‐induced pneumonia, 355.5 (267.3–496.3) U/ml in bacterial pneumonia, 720.5 (466.3–1494) U/ml in COP, 694.5 (384.5–751) U/ml in CVD‐IP, 376 (174–481.5) U/ml in HP, 522 (300–1918) U/ml in AIP, and 307.5 (198–427.8) U/ml in others. The distribution of serum KL‐6 and SP‐D levels in each disease is shown in Figure 3. We built a multivariable logistic regression model to assess the confounding effects of age, sex, and cancer in the relationship between the diagnosis of COVID‐19 pneumonia and serum KL‐6 and SP‐D levels (Table 2). After adjustment for these variables, we found that the increase in serum SP‐D (odds ratio [OR] 0.80, 95% confidence interval [CI]: 0.71–0.90) was negatively associated with the diagnosis of COVID‐19 pneumonia. However, serum KL‐6 levels were not independently associated with COVID‐19 pneumonia diagnosis. Cancer was also negatively associated with the diagnosis of COVID‐19 (OR 0.06, 95% confidence intervals [CI]: 0.01–0.54). Age and sex were not independently associated with the COVID‐19 pneumonia diagnosis.
Figure 2.
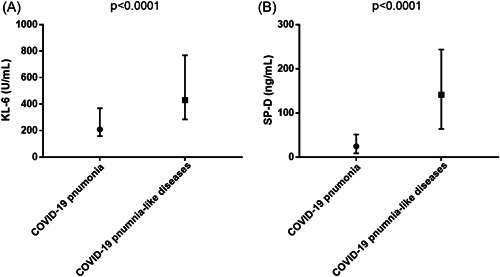
Comparison of serum KL‐6 and SP‐D levels between patients with COVID‐19 pneumonia and COVID‐19 pneumonia‐like diseases. (A) Serum KL‐6 levels of patients with COVID‐19 pneumonia‐like are significantly higher than that of patients with COVID‐19 pneumonia (p < 0.0001). Median (IQR) levels of serum KL‐6 in patients with COVID‐19 pneumonia and COVID‐19 pneumonia‐like diseases were 208.5 (157.5–368.5) U/ml and 430 (284.5–768.5) U/ml, respectively. (B) Serum SP‐D levels of patients with COVID‐19 pneumonia‐like are significantly higher than that of patients with COVID‐19 pneumonia (p < 0.0001). Median (IQR) levels of serum SP‐D in patients with COVID‐19 pneumonia and COVID‐19 pneumonia‐like diseases were 24.7 (8.6–51.0) ng/ml and 141 (63.7–243.5) ng/ml, respectively. COVID‐19, coronavirus disease; IQR, interquartile range; KL‐6, Krebs von den Lungen 6; SP‐D, surfactant protein D
Figure 3.
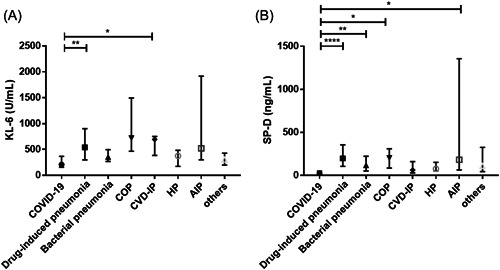
Serum KL‐6 and SP‐D levels of patients with COVID‐19 pneumonia and COVD‐19 pneumonia‐like disease. (A) The serum KL‐6 level of COVID‐19 pneumonia was significantly lower than that of drug‐induced pneumonia or CVD‐IP. The median (IQR) levels of serum KL‐6 were 208.5 (157.5–368.5) U/ml in COVID‐19 pneumonia, 541 (299–904) U/ml in drug‐induced pneumonia, 355.5 (267.3–496.3) U/ml in bacterial pneumonia, 720.5 (466.3–1494) U/ml in COP, 694.5 (384.5–751) U/ml in CVD‐IP, 376 (174–481.5) U/ml in HP, 522 (300–1918) U/ml in AIP, and 307.5 (198–427.8) U/ml in others. (B) The serum SP‐D level of patients with COVD‐19 pneumonia was significantly lower than that of patients with drug‐induced pneumonia or bacterial pneumonia or COP or AIP. The median (IQR) levels of serum SP‐D were 24.7 (8.6–51.0) ng/ml in COVID‐19 pneumonia, 198 (106–356) ng/ml in drug‐induced pneumonia, 119 (51.5–222.5) ng/ml in bacterial pneumonia, 202.5 (86–309.3) ng/ml in COP, 63.7 (29.9–160.3) ng/ml in CVD‐IP, 78.7 (52.6–152.2) ng/ml in HP, 182 (64–1355) ng/ml in AIP, and 113 (44.8–325.8) ng/ml in others. *p < 0.05, ** p < 0.01, ****p < 0.0001. AIP, acute interstitial pneumonia; COP, cryptogenic organizing pneumonia; COVID‐19, coronavirus disease; CVD‐IP, collagen vascular disease‐related interstitial pneumonia; HP, hypersensitivity pneumonitis; IQR, interquartile range; KL‐6, Krebs von den Lungen 6; SP‐D, surfactant protein D
Table 2.
Results from multivariable logistic regression model
| Variables | OR (95% CI) | pValue |
|---|---|---|
| Age (per 10‐year increase) | 0.98 (0.74–1.30) | 0.864 |
| Female | 1.12 (0.41–3.08) | 0.831 |
| KL‐6 (per 100 U/ml increase) | 0.87 (0.70–1.08) | 0.213 |
| SP‐D (per 10 ng/ml increase) | 0.80 (0.71–0.90) | 0.0003 |
| Cancer | 0.06 (0.01–0.54) | 0.0122 |
Note: Significant results (p < 0.05) are shown in bold type. Abbreviations: CI, Confidence interval; KL‐6, Krebs von den Lungen 6; OR, odds ratio; SP‐D, Serum surfactant protein D.
3.3. ROC analysis of serum KL‐6 and SP‐D levels for distinguishing COVID‐19 pneumonia patients from COVID‐19 pneumonia‐like diseases
The areas under the ROC curves (95% CI) of serum KL‐6 and SP‐D levels for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases were 0.761 (0.675–0.847) and 0.874 (0.812–0.936), respectively (Figure 4). The area under the ROC curve of serum SP‐D was significantly larger than that of serum KL‐6 (p = 0.0213), suggesting that the diagnostic performance of serum SP‐D is better than that of serum KL‐6. According to the ROC analysis, the optimal cut‐off level for serum SP‐D was 63.7 ng/ml. When the cut‐off level was applied, the sensitivity and specificity of serum SP‐D were 75.4% and 85.2%, respectively. Even when the ROC analysis was performed in patients without cancer, the areas under the ROC curves (95% CI) of serum KL‐6 and SP‐D levels were 0.750 (0.651–0.849) and 0.881 (0.8150–0.9463), respectively, suggesting that the diagnostic performances of serum SP‐D and KL‐6 were almost the same between the population including cancer and not including cancer.
Figure 4.
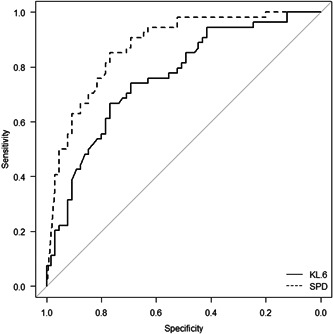
The ROC curves of serum KL‐6 and SP‐D levels for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases. Area under the ROC curve of serum SP‐D level was 0.874, while that of serum KL‐6 level was 0.761. COVID‐19, coronavirus disease; KL‐6, Krebs von den Lungen 6; ROC, receiver operating characteristic; SP‐D, surfactant protein D
4. DISCUSSION
Our findings show that the levels of serum KL‐6 and SP‐D in patients with COVID‐19 pneumonia were significantly lower than those in patients with COVID‐19 pneumonia‐like diseases, suggesting that serum KL‐6 and SP‐D are useful for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases. In addition, according to ROC analysis, serum SP‐D can distinguish patients more efficiently than serum KL‐6, and the diagnostic performance of serum SP‐D could be sufficient for use in clinical practice.
KL‐6 and SP‐D are both secreted by type II pneumocytes. The role of both molecules has been investigated in ILD because the common pathological features of ILD include type II pneumocyte injury or remodeling. 14 KL‐6 is a mucinous glycoprotein with a high molecular weight. The production of KL‐6 is upregulated in regenerating type II pneumocytes and serum KL‐6 has been reported to be useful as a diagnostic and prognostic biomarker in ILD. 14 SP‐D is a multimeric collectin that exhibits anti‐inflammatory activity and increases opsonization. 15 The concentration of serum SP‐D increases as a result of translocation from the alveoli to the bloodstream caused by the impairment of the alveolar‐capillary membrane. 16 Thus, the increase in serum SP‐D is thought to reflect protein translocation caused by lung injury and the clinical implication of serum SP‐D is also evaluated in inflammatory disorders of the lung, including ILD. 7 , 17 SARS‐CoV‐2 primarily infects Type II pneumocytes. 18 Infection with the virus is followed by cell death, resulting in lung injury. 19 The clinical implications of serum KL‐6 and SP‐D levels have been investigated in the above‐stated context. As per reports, both serum KL‐6 and SP‐D levels are elevated in patients with severe COVID‐19 and, therefore, these constituents are useful biomarkers of disease severity. 9 , 11 , 13 , 20 , 21 In addition to disease severity, serum KL‐6 was reported to predict pulmonary fibrosis followed by infection with SARS‐CoV‐2 10 , 22 and serum SP‐D was reported to be useful for distinguishing pandemic influenza A (H1N1) from COVID‐19 in patients who required mechanical ventilation. 23 Given the previous reports demonstrating the relationship between disease severity and serum KL‐6 and SP‐D levels, the proportion of patients with severe COVID‐19 may influence the diagnostic performance of serum KL‐6 and SP‐D. In this study, 20% of COVID‐19 patients presented with respiratory failure. According to an early report, 81% of COVID‐19 patients did not present with respiratory failure. 24 Thus, the population of enrolled patients with COVID‐19 in this study was considered similar to the general population of patients with COVID‐19. Therefore, we believe that our results can be applied in clinical practice.
In this study, the diagnostic performance of serum SP‐D was superior to that of serum KL‐6. A possible explanation for the superior performance of serum SP‐D is that serum SP‐D level was greater in bacterial pneumonia than in COVID‐19 pneumonia, while serum KL‐6 level was not elevated in bacterial pneumonia. This finding is consistent with a previous report. 7 Although the reason for the difference between serum KL‐6 and SP‐D levels in bacterial pneumonia is unknown, the difference in the mechanisms of elevation of both molecules in the blood may affect the difference between the serum levels. Bacterial pneumonia might disrupt the alveolar‐capillary membrane, but it cannot cause regeneration of type II pneumocytes, at least in the early stages of the illness. Another conceivable explanation is that the production of SP‐D from type II pneumocytes might be impaired by the destruction of type II pneumocytes followed by an invasion of SARS‐CoV‐2 because the serum SP‐D level of patients with COVID‐19 pneumonia was very low. Although the median serum SP‐D level of patients with COVID‐19 was 24.7 (8.6–51.0) ng/ml in this study, the cut‐off level of serum SP‐D for distinguishing interstitial pneumonia from healthy controls is generally 110 ng/ml and the mean serum SP‐D level of healthy volunteers is reported to be 40–98 ng/ml. 17 To clarify this point, further investigations involving healthy volunteers are required.
KL‐6 was originally discovered as a soluble tumor‐associated antigen which can be detected in sera and effusions in patients with cancer 25 and several reports have addressed the relationship between serum KL‐6 levels and cancer. Serum KL‐6 is reported to be useful as a tumor marker in several types of cancers. 26 , 27 , 28 Several reports have investigated the pathological role of SP‐D in cancer. 29 , 30 However, the clinical implications of serum SP‐D are poorly understood. In our cohort, the proportion of patients with cancer was higher in patients with COVID‐19 pneumonia‐like disease, because most cases of drug‐induced pneumonia were caused by antitumor drugs. According to the multivariable logistic regression analysis, in which cancer was used as an independent variable, serum KL‐6 was not independently associated with the diagnosis of COVID‐19 pneumonia, while serum SP‐D was independently associated with the diagnosis of COVID‐19 pneumonia. Even though the serum KL‐6 level of COVID‐19 pneumonia was significantly lower in the population without cancer in this study, serum KL‐6 levels can be affected by cancer. In this respect, serum SP‐D is superior to serum KL‐6 for the diagnosis of COVID‐19 pneumonia.
This study had several limitations. First, this was a retrospective, single‐center study, and the number of enrolled patients was small. In particular, only a few patients with diseases such as COP, CVD‐IP, HP, and AIP were enrolled; therefore, the data for these diseases was insufficient. Indeed, serum SP‐D levels have been reported to increase in HP and CVD‐IP. 17 However, serum SP‐D levels in patients with HP and CVD‐IP were not greater than those in patients with COVID‐19 pneumonia in this study. Second, the distribution of causes of COVID‐19 pneumonia‐like disease can differ depending on the epidemiological pattern of diseases and the role of hospitals in the local community. Therefore, the average serum SP‐D and KL‐6 levels in COVID‐19 pneumonia‐like diseases may differ depending on the local situation. However, we believe that it is useful for the diagnosis of COVID‐19 pneumonia in any situation to recognize that the serum SP‐D level was low in most patients with COVID‐19 pneumonia. Third, our findings were based on the data of patients infected with wild‐type SARS‐CoV‐2. Numerous variants have continued to emerge and the clinical features have been changing based on the variants. Thus, the utility of serum SP‐D also can change depending on the variants.
The management of false‐negative NAAT results and antigen testing is a major challenge in the clinical setting, especially for patients who require hospitalization. Although chest CT is useful for the prevention of missed diagnosis of COVID‐19 pneumonia, it has the disadvantage of low specificity. This study shows that serum SP‐D might compensate for the disadvantages of chest CT and can be potentially useful in managing patients with negative NAAT results for SARS‐CoV‐2 and abnormal chest shadows mimicking COVID‐19 pneumonia. Therefore, serum SP‐D is a promising biomarker for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases.
AUTHOR CONTRIBUTIONS
Yuki Togashi: investigation; writing – original draft. Yuta Kono: conceptualization; formal analysis; methodology; project administration; writing – review & editing. Takashi Okuma: data curation. Nao Shioiri: data curation. Reimi Mizushima: data curation. Akane Tanaka: data curation. Mayuko Ishiwari: data curation. Kazutoshi Toriyama: data curation. Ryota Kikuchi: data curation. Hiroyuki Takoi: data curation. Shinji Abe: project administration; supervision.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
TRANSPERENCY STATEMENT
The corresponding author declares that this manuscript is honest, precise and transparent account of the study. Any aspects of the study have not been omitted.
ETHICS STATEMENT
The study was a retrospective study, and it was approved by the Ethics Committee of the Tokyo Medical University Hospital (approval number, T2021‐0183).
ACKNOWLEDGMENTS
None. This study did not receive funding from any sources.
Togashi Y, Kono Y, Okuma T, et al. Surfactant protein D: a useful biomarker for distinguishing COVID‐19 pneumonia from COVID‐19 pneumonia‐like diseases. Health Sci. Rep. 2022;5:e622. 10.1002/hsr2.622
DATA AVAILABILITY STATEMENT
The data analyzed in this study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Weissleder R, Lee H, Ko J, Pittet MJ. COVID‐19 diagnostics in context. Sci Transl Med. 2020;12(546):eabc1931. [DOI] [PubMed] [Google Scholar]
- 2. Dinnes J, Deeks JJ, Berhane S, et al. Rapid, point‐of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database Syst Rev. 2021;3(3):CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson KD, Harris C, Cain JK, Hummer C, Goyal H, Perisetti A. Pulmonary and extra‐pulmonary clinical manifestations of COVID‐19. Front Med (Lausanne). 2020;7:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alsharif W, Qurashi A. Effectiveness of COVID‐19 diagnosis and management tools: a review. Radiography (Lond). 2021;27(2):682‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ojha V, Mani A, Pandey NN, Sharma S, Kumar S. CT in coronavirus disease 2019 (COVID‐19): a systematic review of chest CT findings in 4410 adult patients. Eur Radiol. 2020;11:6129‐6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlicchi E, Gemma P, Poerio A, Caminati A, Vanzulli A, Zompatori M. Chest‐CT mimics of COVID‐19 pneumonia—a review article. Emerg Radiol. 2021;28(3):507‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohnishi H, Yokoyama A, Kondo K, et al. Comparative study of KL‐6, surfactant protein‐A, surfactant protein‐D, and monocyte chemoattractant protein‐1 as serum markers for interstitial lung diseases. Am J Respir Crit Care Med. 2002;165(3):378‐381. [DOI] [PubMed] [Google Scholar]
- 8. Yamakawa H, Hagiwara E, Ikeda S, et al. Evaluation of changes in the serum levels of Krebs von den Lungen‐6 and surfactant protein‐D over time as important biomarkers in idiopathic fibrotic nonspecific interstitial pneumonia. Respir Investig. 2019;57(5):422‐429. [DOI] [PubMed] [Google Scholar]
- 9. Deng K, Fan Q, Yang Y, et al. Prognostic roles of KL‐6 in disease severity and lung injury in COVID‐19 patients: a longitudinal retrospective analysis. J Med Virol. 2021;93(4):2505‐2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. d'Alessandro M, Bergantini L, Cameli P, et al. Serial KL‐6 measurements in COVID‐19 patients. Intern Emerg Med. 2021;16(6):1541‐1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. d'Alessandro M, Cameli P, Refini RM, et al. Serum KL‐6 concentrations as a novel biomarker of severe COVID‐19. J Med Virol. 2020;92(10):2216‐2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghati A, Dam P, Tasdemir D, et al. Exogenous pulmonary surfactant: a review focused on adjunctive therapy for severe acute respiratory syndrome coronavirus 2 including SP‐A and SP‐D as added clinical marker. Curr Opin Colloid Interface Sci. 2021;51:101413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alay H, Laloglu E. The role of angiopoietin‐2 and surfactant protein‐D levels in SARS‐CoV‐2‐related lung injury: a prospective, observational, cohort study. J Med Virol. 2021;93:6008‐6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ishikawa N, Hattori N, Yokoyama A, Kohno N. Utility of KL‐6/MUC1 in the clinical management of interstitial lung diseases. Respir Investig. 2012;50(1):3‐13. [DOI] [PubMed] [Google Scholar]
- 15. Crouch EC. Surfactant protein‐D and pulmonary host defense. Respir Res. 2000;1(2):93‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang L, Ikegami M, Korfhagen TR, et al. Neither SP‐A nor NH2‐terminal domains of SP‐A can substitute for SP‐D in regulation of alveolar homeostasis. Am J Physiol Lung Cell Mol Physiol. 2006;291(2):L181‐L190. [DOI] [PubMed] [Google Scholar]
- 17. Hartl D, Griese M. Surfactant protein D in human lung diseases. Eur J Clin Invest. 2006;36(6):423‐435. [DOI] [PubMed] [Google Scholar]
- 18. Wahl A, Gralinski LE, Johnson CE, et al. SARS‐CoV‐2 infection is effectively treated and prevented by EIDD‐2801. Nature. 2021;591(7850):451‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mason RJ. Pathogenesis of COVID‐19 from a cell biology perspective. Eur Respir J. 2020;55(4):2000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamaya T, Hagiwara E, Baba T, et al. Serum Krebs von den Lungen‐6 levels are associated with mortality and severity in patients with coronavirus disease 2019. Respir Investig. 2021;59(5):596‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scotto R, Pinchera B, Perna F, et al. Serum KL‐6 could represent a reliable indicator of unfavourable outcome in patients with COVID‐19 pneumonia. Int J Environ Res Public Health. 2021;18(4):2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xue M, Zhang T, Chen H, et al. Krebs Von den Lungen‐6 as a predictive indicator for the risk of secondary pulmonary fibrosis and its reversibility in COVID‐19 patients. Int J Biol Sci. 2021;17(6):1565‐1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choreño‐Parra JA, Jiménez‐Álvarez LA, Ramírez‐Martínez G, et al. Expression of surfactant protein D distinguishes severe pandemic influenza A (H1N1) from Coronavirus Disease 2019. J Infect Dis. 2021;224(1):21‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Z, McGoogan JM. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID‐19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239‐1242. [DOI] [PubMed] [Google Scholar]
- 25. Kohno N, Akiyama M, Kyoizumi S, Hakoda M, Kobuke K, Yamakido M. Detection of soluble tumor‐associated antigens in sera and effusions using novel monoclonal antibodies, KL‐3 and KL‐6, against lung adenocarcinoma. Jpn J Clin Oncol. 1988;18(3):203‐216. [PubMed] [Google Scholar]
- 26. Tomita M, Ayabe T, Chosa E, Nose N, Nakamura K. Prognostic significance of a tumor marker index based on preoperative serum carcinoembryonic antigen and Krebs von den Lungen‐6 levels in non‐small cell lung cancer. Asian Pac J Cancer Prev. 2017;18(1):287‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogawa Y, Ishikawa T, Ikeda K, et al. Evaluation of serum KL‐6, a mucin‐like glycoprotein, as a tumor marker for breast cancer. Clin Cancer Res. 2000;6(10):4069‐4072. [PubMed] [Google Scholar]
- 28. Sato S, Kato T, Abe K, et al. Pre‐operative evaluation of circulating KL‐6 levels as a biomarker for epithelial ovarian carcinoma and its correlation with tumor MUC1 expression. Oncol Lett. 2017;14(1):776‐786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mangogna A, Belmonte B, Agostinis C, et al. Pathological significance and prognostic value of surfactant protein D in cancer. Front Immunol. 2018;9:1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar J, Murugaiah V, Sotiriadis G, et al. Surfactant protein D as a potential biomarker and therapeutic target in ovarian cancer. Front Oncol. 2019;9:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data analyzed in this study are available from the corresponding author on reasonable request.


