Figure 2.
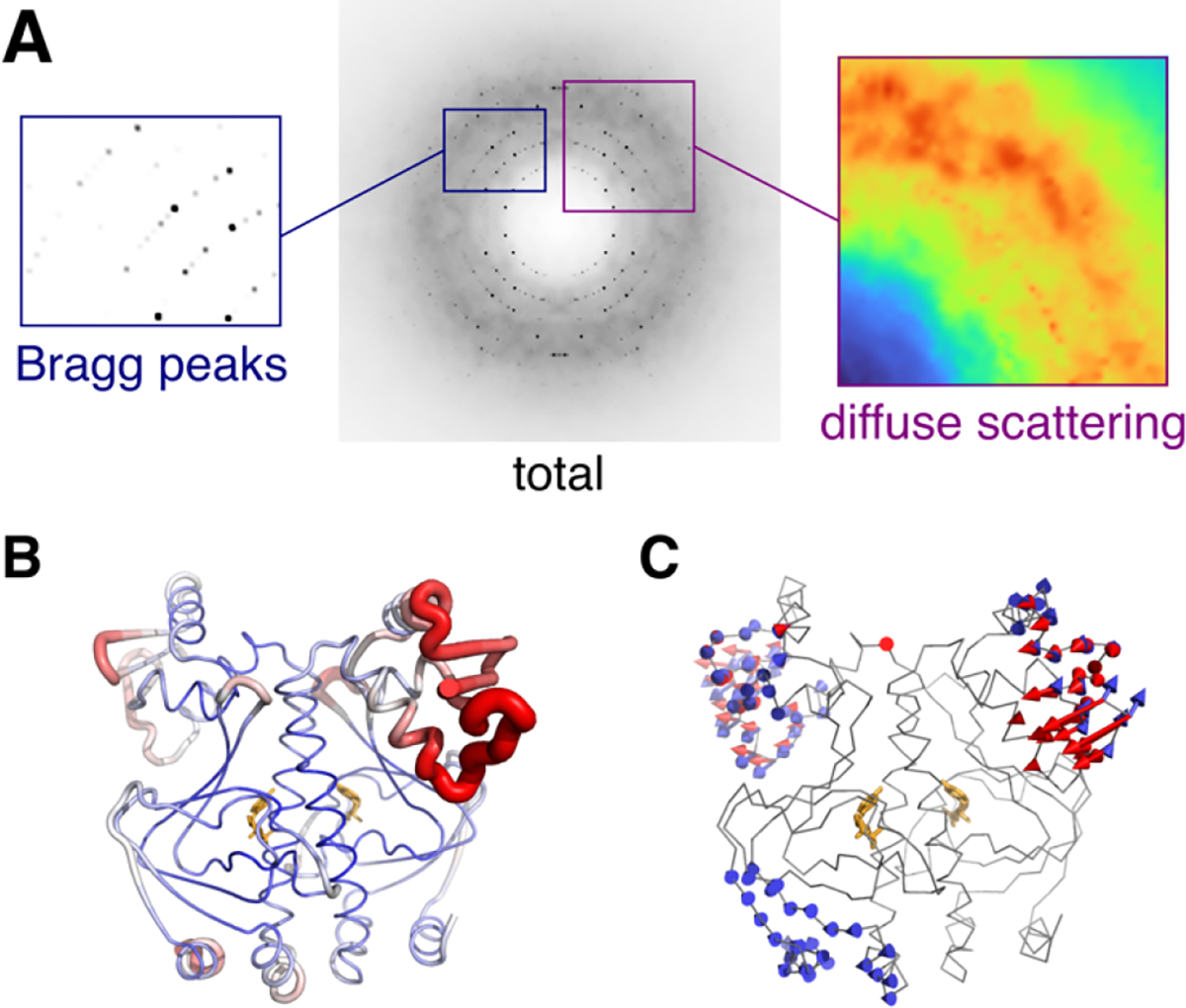
Components of total scattering illustrated using the experimental X-ray structure of CAP (PDB: 1g6n53) with dynamics added using ENM simulations54 of one unit cell. (A) The simulated diffraction image contains two signals: Bragg peaks (left) that depend on the average structure, and diffuse scattering (right) that arises from correlated atomic displacements (in this case, vibrations of the ENM). (B) B-factors refined to experimental Bragg data vary along the polypeptide chain (blue to red). (C) Normal modes of the ENM seem to explain regions of high experimental B-factor. The reality of such collective motions can be verified by diffuse scattering analysis.
