Figure 3.
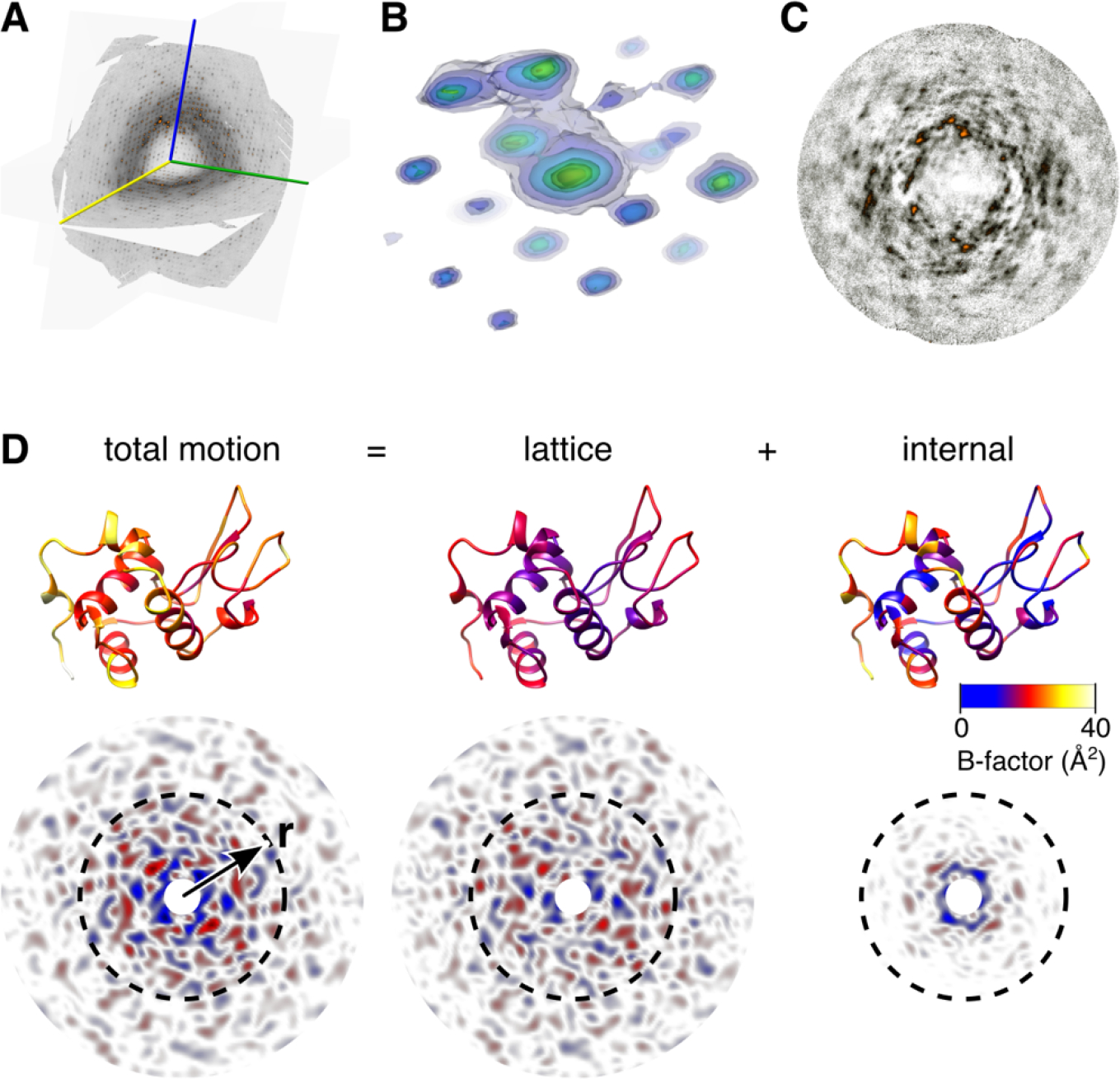
Total scattering analysis separates motion into lattice and internal components54. (A) Three-dimensional map of diffuse scattering from triclinic lysozyme shown as intersecting central slices. A movie showing this volume from multiple perspectives is available online. The scattering includes an intense isotropic ring that may be subtracted to better visualize the halo and cloudy features. (B) Around Bragg peaks are three-dimensional halos (shown here as transparent contours, blue to yellow) attributed to thermally excited lattice vibrations. (C) Cloudy features due to short-ranged correlated motion are most visible in sections mid-way between Bragg planes. (D) Total motion and correlations are quantified using B-factor (top, PDB 6o2h54) and diffuse Patterson maps (bottom), which report electron density fluctuations vs. inter-atomic vector, r. A lattice dynamics model fit to diffuse halos accounts for most of the B-factor for well-ordered atoms (total vs. lattice, top) and the correlated motions at large distances (total vs. lattice, bottom), but underestimates those at short distance (r < 10 Å, dashed circles). An ENM describing protein dynamics was fit to the residual B-factors (top right). The simulated diffuse Patterson of the protein dynamics model (bottom right) explains the remaining short-ranged correlations.
