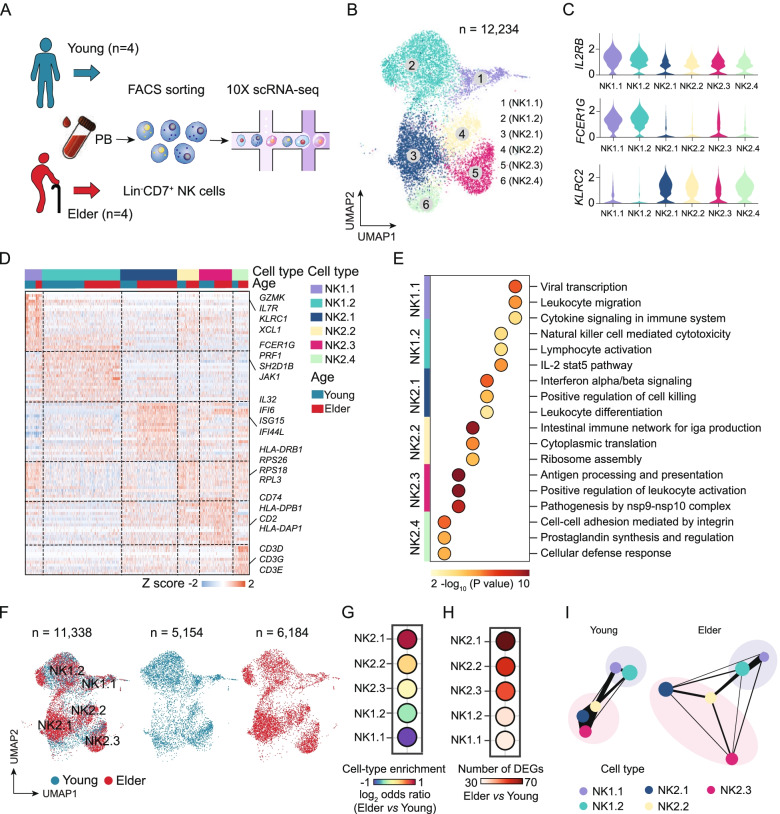
Fig. 2.
ScRNA-seq analysis of aged human blood NK cells reveals age-associated alterations in memory-like NK cells. A Peripheral NK cells (Lin−CD7+) were sorted from young and elderly individuals and analysed with the 10X Genomics single-cell sequencing platform. B UMAP projections of 12,234 NK cells. The different colours represent the 6 NK cell subpopulations (left). Each dot represents a single cell. C, Violin plot showing the gene expression of IL2RB, FCER1G, and KLRC2 in each cell subpopulation. D Heatmap showing the differentially expressed genes among the NK cell subpopulations from young and elderly individuals. E GO enrichment analysis showing the terms of the differentially expressed genes for the indicated NK cell subpopulations from young and elderly individuals. The colour indicates the -log10 (P-value) enrichment for each GO term. F UMAP projections of 11,338 NK cells, with 5154 from young individuals (middle) and 6184 cells from elderly individuals (right). Each dot represents a single cell. G Dot plot showing the proportions of the NK cell subpopulations in an elder vs. young comparison. H Dot plot showing a comparison of the number of differentially expressed genes in the indicated NK cell subpopulations in an elderly vs. young comparison. I PAGA graph showing the connectivity between the NK subgroups in the young and elderly groups. Each of the coloured nodes is an NK subpopulation, and the node size is proportional to the number of cells in the subpopulation. The thickness of the edge shows the strength of the connectivity between subpopulations. DEGs, differentially expressed genes