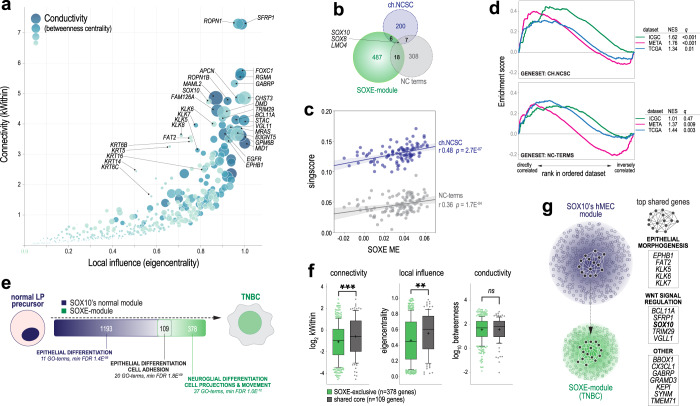
Fig. 4. The SOXE-module drives the transition from normal mammary epithelial stem/progenitor to NCSC-like phenotypic states.
a Influence of SOXE-module genes over network architecture and information flow. kWithin: intramodular ‘connectivity’ based on weighted correlations with all other module genes; Eigencentrality: considers the connectivity of each node’s nearest neighbours as an indicator of ‘local influence’; Betweenness centrality: ‘conductivity’ based on each node’s position along the shortest paths between other nodes (genes with high betweenness are information conduits). Key hub genes are indicated (see Supplementary Table 10 for the full dataset). b Chick (ch.)NCSC and neural crest (NC) terms genesets are largely independent of each other and from the SOXE-module. c Correlations between SOXE-ME values and NCSC genesets (singscore values) in TNBC (n = 106 TCGA cases with tumour cellularity ≥0.6). Correlation coefficients (r) and p values are shown. d GSEA using three TNBC gene expression datasets (ICGC, METABRIC, TCGA). Normalised enrichment scores (NES) and corrected p values (q) shown. e Overlap between members of the SOXE-module and SOX10’s normal breast module (from de novo module identification on n = 97 TCGA normal breast samples; Supplementary Table 12). Generic ontology enrichment results are summarised (full GO term lists in Supplementary Table 13). f Comparison of network structure and information flow metrics (as for (a)) between shared and SOXE-module-exclusive genes. Groups were compared using Mann–Whitney tests (**p = 2.4E-03; ***p = 5.6E-04). Boxes show the 10–90th percentiles and median, with whiskers extending to the minimum and maximum values. Mean is indicated with ‘+’. g Model depicting the mammary epithelial progenitor gene regulatory network core being sustained through transformation and rewired as the SOXE-module in TNBC. Shared hub genes are listed.