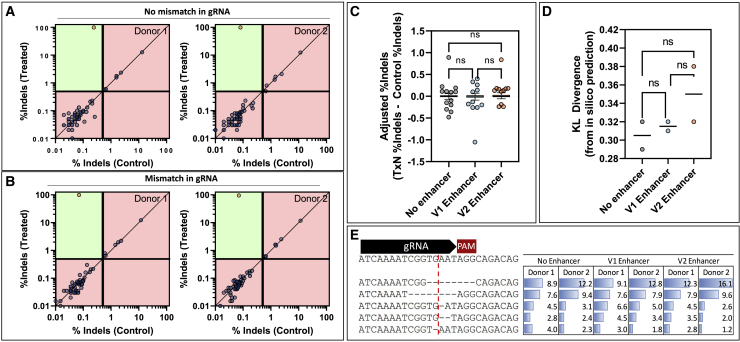
Figure 7.
Characterization of off-target editing and effects of DNA-repair modulators
Editing was quantified using CRISPAltRations at all nominated on- or off-target loci for two healthy donors with paired treatment or controls after being edited with the TRAC guide (A) without a mismatch in the gRNA or (B) with a mismatch in the gRNA. Sites were classified as edited (orange circle) or not edited (blue circle) using a thresholded Fisher’s exact test (p < 0.05) with classification limitations. Green areas indicate areas capable of classification, whereas red areas indicate areas outside classification limits (indels in treatment <0.5%; indels in control >0.4%; <5,000 reads). Using the gRNA with no mismatch, the effects of HDR enhancing small molecules on (C) off-target editing and (D) on-target indel profiles, measured as KL divergence from FORECasT predictions, were investigated (one-way ANOVA with post-hoc Tukey’s correction; p < 0.05). (E) The contributions of the top five indels for the no enhancer sample are shown for each donor and treatment (cut site shown as red line).