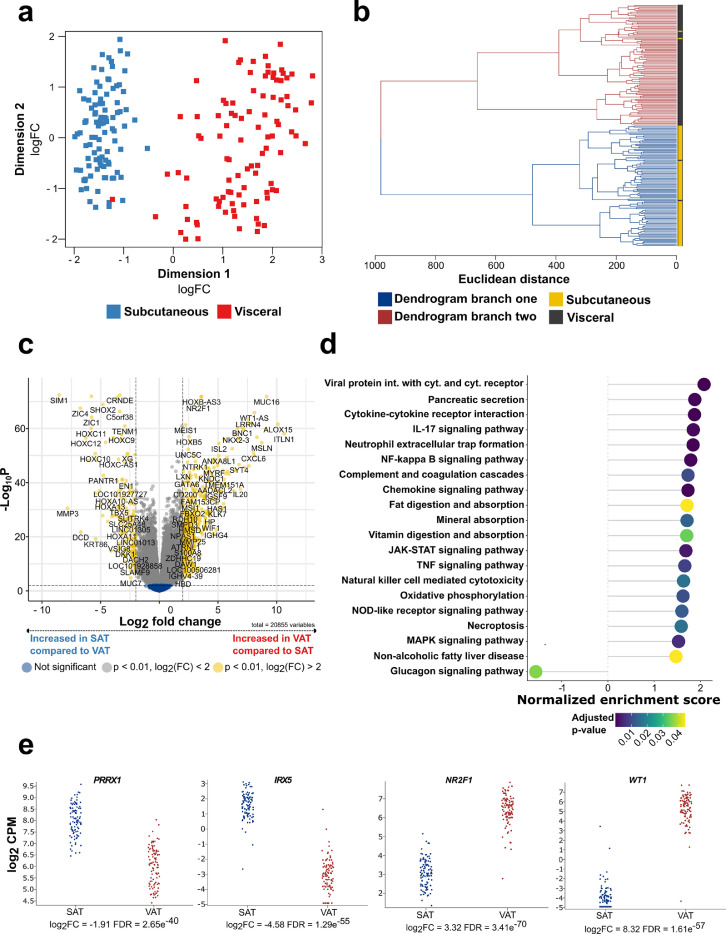
Figure 1.
SAT and VAT are characterized by different transcriptomic signatures
(a) MDS plot displaying the first two dimensions of the data (n = 99). (b) Hierarchical clustering of SAT and VAT samples based on euclidean sample distances (n = 99). (c) Volcano plot showing differentially expressed genes between SAT and VAT (n = 99). (d) Dotplot displaying the top 20 enriched KEGG pathways with an adjusted p-value < 0.05 from gene set enrichment analysis, thereby comparing the expression of KEGG pathway associated genes from SAT and to VAT depots (n =99). Positive scores indicate enrichment of gene-sets in VAT while negative scores indicate enrichment in SAT. (e) Log2 counts per million (CPM) of SAT and VAT lineage specific marker genes (n = 99).