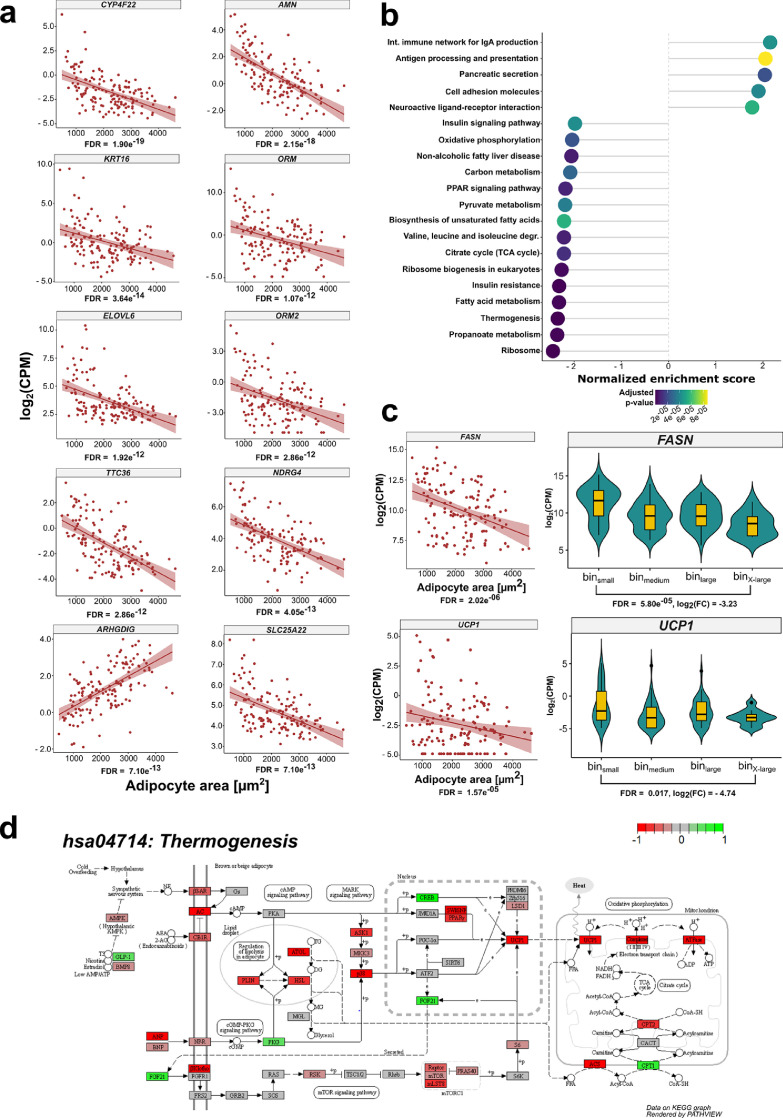
Figure 3.
Adipocyte area is related to gene expression in VAT
(a) Scatterplots displaying the results of the continuous model-based analysis that was applied to GTEx RNA-Seq data from VAT. Log2 counts per million (CPM) are plotted against adipocyte area for 141 individuals. FDR is given for each gene below the plot. Displayed is the relationship between adipocyte area and gene expression for 10 exemplary genes that were ranked highest in the analysis according to their FDR. A regression line including the standard error in a lighter shade of red was added to the plots to visualize the relationship between gene expression and adipocyte area. (b) Dotplot displaying the top 20 enriched KEGG pathways with an adjusted p-value < 0.05 from gene set enrichment analysis in VAT (n = 141). The “Human Diseases” category of the KEGG PATHWAY database is not displayed on the graph. (c) Scatterplot and violin plot assessing the relationship between the expression of FASN, UCP1 and adipocyte area (nscatter = 141, nbin small = 41, nbin medium = 49, nbin large = 40, nbin xlarge = 11). (d) KEGG Pathview graph depicting the different complexes and genes involved in the canonical thermogenesis pathway (hsa04714:Thermogenesis). Red (- 1) indicates thermogenesis related genes that are underrepresented in VAT from the binX-Large (n = 11) group compared to the binsmall (n = 40) group. In contrast, green (+1) displays thermogenesis genes that are overrepresented in VAT from the binX-large group in relation to the binsmall samples. Genes depicted in grey (0) did not show differences between the two groups.