Abstract
The emergence of new SARS-CoV-2 Omicron variant of concern (OV) has exacerbated the COVID-19 pandemic because of a large number of mutations in the spike protein, particularly in the receptor-binding domain (RBD), resulting in highly contagious and/or vaccine-resistant strains. Herein, we present a systematic analysis based on detailed molecular dynamics (MD) simulations in order to understand how the OV RBD mutations affect the ACE2 binding. We show that the OV RBD binds to ACE2 more efficiently and tightly predominantly because of strong electrostatic interactions, thereby promoting increased infectivity and transmissibility compared to other strains. Some of the OV RBD mutations are predicted to affect the antibody neutralization either through their role in the S-protein conformational changes, such as S371L, S373P, and S375F, or through changing its surface charge distribution, such as G339D, N440K, T478K, and E484A. Other mutations, such as K417N, G446S, and Y505H, decrease the ACE2 binding, whereas S447N, Q493R, G496S, Q498R, and N501Y tend to increase it.
Since the outbreak of the COVID-19 pandemic, its pathogen SARS-CoV-2 has continuously mutated and evolved, resulting in the emergence of major variants of concern (VOC). These VOC have been observed to alter the virus characteristics, such as infectivity, transmissibility, antigenicity, and pathogenicity.1 The most recently identified SARS-CoV-2 VOC is the Omicron variant (OV) (B.1.1.529), which has quickly become the dominant strain.2,3 It has the highest number of amino acid (AA) mutations of any known SARS-CoV-2 VOC, with over 30 mutations in the spike (S) protein, of which 15 are in the receptor-binding domain (RBD),4 the main target for vaccine and treatment developments.5−8 The presence of many mutations in OV S-protein has raised concerns about elevated transmissibility, immunological escape, and vaccine and treatment failures.9−15 OV has been identified to contain several key mutations observed also in other SARS-CoV-2 VOC, such as K417N, E484A, and N501Y, that change the virus sensitivity to neutralization or increase the infectivity.16 Moreover, it contains many novel mutations that have not been observed previously, and their biological effects are largely unknown. Because the binding between RBD and the angiotensin-converting enzyme-2 (ACE2) facilitates viral entry, initiating the infection process, the fundamental understanding of how the OV RBD interacts with ACE2 is pivotal for understanding the viral infection mechanism and its evolution, as well as for therapeutic development of effective means to reduce its spread.
The present study aims to investigate how OV mutations affect the binding strength between RBD and ACE2 by highlighting the role of each mutation, its underlying mechanism, and the pertinent binding driving forces. All-atom molecular dynamics (MD) simulations in explicit solvent have been implemented in order to study the dynamics and binding mechanism of the RBD-ACE2 system, followed by the molecular mechanics (MM) generalized Born surface area (MM-GBSA) method to predict the binding affinity and the binding profile. The results are compared with the previously reported analysis of the unmutated, wild type (WT) RBD-ACE2 system17 in order to assess the effect of each mutation and the nature of its interaction based on the per-residue and pairwise decomposition schemes. MD simulations have proven valuable to investigate the dynamic and binding processes of the RBD-ACE2 complex in WT and many previous VOC.18−25
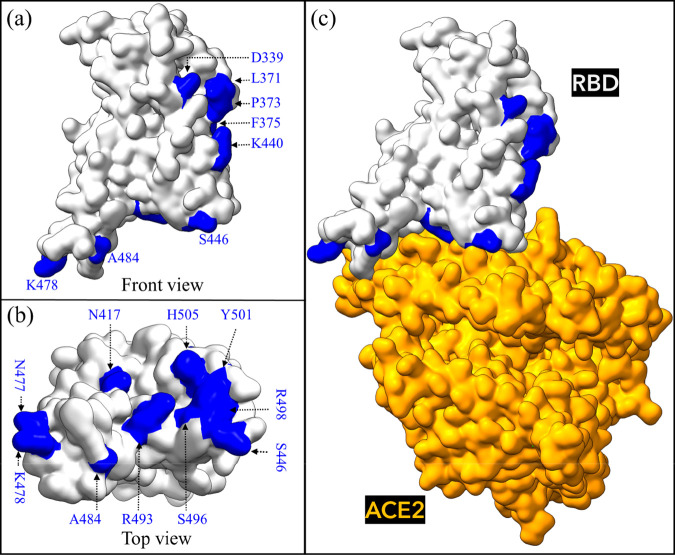
We implement a procedure, previously developed for both Alpha and Beta VOC, to build a computational model of the Omicron RBD-ACE2 system.17 We briefly summarize it as follows. First, the interface structure of RBD-ACE2 complex (PBD ID:6M0J) is used as a template to create the OV RBD-ACE2 system, including all 15 RBD mutations:26 G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H.4 The last 10 mutations are subsequently exposed to the ACE2 receptor as shown in Figure 1.
Figure 1.
Omicron RBD-ACE2 interface system. (a) Front view of unbound Omicron variant (OV) RBD with all 15 mutations shown in blue and (b) top view. (c) Bound OV RBD-ACE2 model.
To substitute the AAs of WT with mutated AAs of OV, we used the Dunbrack backbone-dependent rotamer library27 implemented by UCSF Chimera,28 which also allows for a careful control of backbone dihedral angles. For example, the torsion angles of K417N and N501Y mutations from the available RBD Beta VOC have been used in our OV model,29 as well as the angles of T478K mutation in the Delta variant.30 Second, the resulting OV RBD-ACE2 complex is solvated with 27000 explicit water molecules together with the appropriate number of ions (1 Zn2+, 1 Cl–, and 22 Na+) using the TIP3P explicit water model, implemented in AMBER,31,32 with the most recent ff14SB force field used for parametrizations of intermolecular and intramolecular interactions in the OV complex.33 Third, the same MD steps of minimization, heating, equilibration, and two MD production runs were reproduced, but this time the MD runs were extended over 500 ns (1 μs in total).17 For each run, 2500 frames are extracted from the entire simulation and used for the binding free energy (BFE) postprocess analysis. Finally, the MM-GBSA method17,34−37 was applied to compute the BFE and to quantify the complete binding profile,17 allowing for per-residue and pairwise BFE decompositions to identify the role and the nature of interaction for each mutation in the OV model.
Figure 2a shows the root-mean-square deviation (RMSD) and their frequency distributions for two replicate MD simulations of the OV. Overall, the complex achieves stable interfacial interactions as shown by small RMSD fluctuations. We find that the OV RBD-ACE2 complex has a relatively RMSD smaller than that previously reported of the WT from 100 ns MD simulations (average from both runs is 2.32 vs 2.53 Å of WT).17 The 15 OV RBD mutations thus lead to a more stable interfacial complex with ACE2. Surprisingly, the inset figure depicts a higher overlap between two MD runs. Even though the simulation time is only 500 ns, this is a good sign that our simulations are reproducible and convergent. This is even more pronounced by the value of the root mean squared inner product (RMSIP) of 0.82 for the first two principal components (PC1 and PC2) (Figure S1 in the Supporting Information). This behavior is also evident from time evolutions and distribution of BFE (Figure S2).
Figure 2.
Stability of the OV RBD-ACE2 complex and slight conformation changes of mutated residues at RBD core. (a) Time course and distribution of the root mean square deviation (RMSD) of the heavy atoms of OV RBD-ACE2 complex in both MD runs. (b) RMSF for 15 residues that are mutated in OV and compared with WT.
In comparison with WT, the OV mutations are seen to only slightly change the RBD structure. This is even more pronounced when their root-mean-square fluctuations (RMSFs) are compared, as shown in Figure S3. The average RMSF of mutated RBD in OV is 1.49 Å vs 1.29 Å of WT RBD, indicating that the OV RBD is relatively less rigid than WT RBD, thus allowing for larger fluctuations. Figure 2b compares the RMSF of only the mutated AAs in OV vs WT. In particular, D339, L371, P373, and F375 are relatively more flexible than their WT counterparts. This is consistent with a recent study based on the cryo-EM S-protein structure, showing that the L371, P373, and F375 significantly alter the conformation and mobility of RBD,12 thus demonstrating that the computational MD simulations can confirm and reproduce what is observed experimentally.
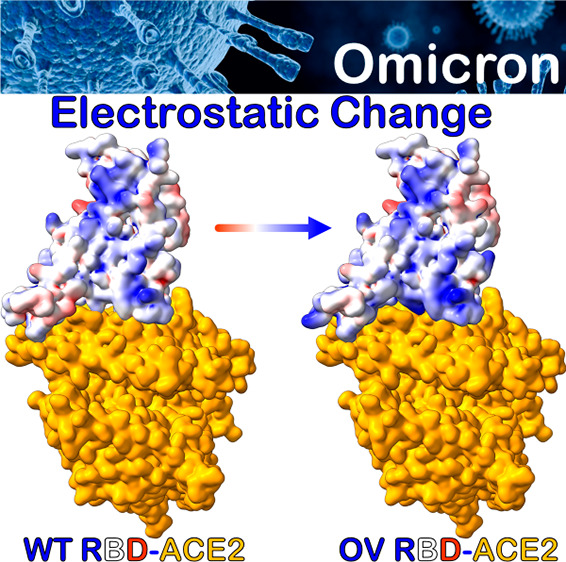
The MM-GBSA method has been applied to calculate the BFE of the OV RBD-ACE2 system at 310 K (37 °C), with neutral pH (7.4) and 0.15 M uniunivalent NaCl salt concentration. Table 1 presents the BFE and its decomposition for OV and in comparison with WT.17 It shows that the OV RBD binds ACE2 more strongly than WT, with relative binding energy of −1.67 kcal/mol, consistent with recent experimental and computational studies.11−13,38−40 Interestingly, our predicted BFE value of −14.53 kcal/mol for OV is close to the Alpha BFE value of −14.7 kcal/mol.17 The complete thermodynamic decomposition listed in Table 1 shows the Coulomb electrostatic interaction (ΔEele) of OV to be more than twice that of WT. The increase in ΔEele of OV is mainly the result of five AAs at the RBM changing from polar to positively charged residues (N440K, T478K, Q493R, Q498R, and Y505H). The electrostatic component is thus seen as the main reason behind the more effective binding of ACE2 and OV RBD, which may also elucidate the reason behind the highly contagious nature of OV (Figure S4). The electrostatic interaction has been shown to be the primary source of increasing the binding of SARS-CoV-2 to ACE2 compared to SARS-CoV as well as enhancing the binding affinity of VOC to ACE2.23−25 ΔEele of OV, however, creates a higher desolvation energy (ΔGGB) that is indispensable in the formation process and cannot be avoided (Table 1). On the other hand, the van der Waals interaction (ΔEvdW) plays a key role in stabilizing and governing the RBD–ACE2 interaction as well as enhancing their binding by gaining −2.71 kcal/mol as compared to WT.
Table 1. Decomposition of BFE (kcal·mol–1) of RBD-ACE2 Complex in OV and WT.
| energy | OV (SEM) | WT (SEM)a | ΔΔGb |
|---|---|---|---|
| ΔEvdW | –92.81 (0.1) | –90.10 (0.1) | –2.71 |
| ΔEele | –1486.5 (0.65) | –700.92 (0.6) | –785.58 |
| ΔEMM | –1579.31 (0.66) | –791.03 (0.7) | –788.28 |
| ΔGGB | 1534.19 (0.64) | 748.49 (0.6) | 785.7 |
| ΔGSA | –13.27 (0.01) | –13.21 (0) | –0.06 |
| ΔGsol | 1520.92 (0.64) | 735.28 (0.6) | 785.64 |
| –TΔS | 43.86 | 42.89 | 0.97 |
| ΔGbind | –14.53 (0.1) | –12.86 (0.1) | –1.67 |
All values are taken from ref (17), and SEM is the standard error of the mean.
ΔΔG = ΔGOV – ΔGWT.
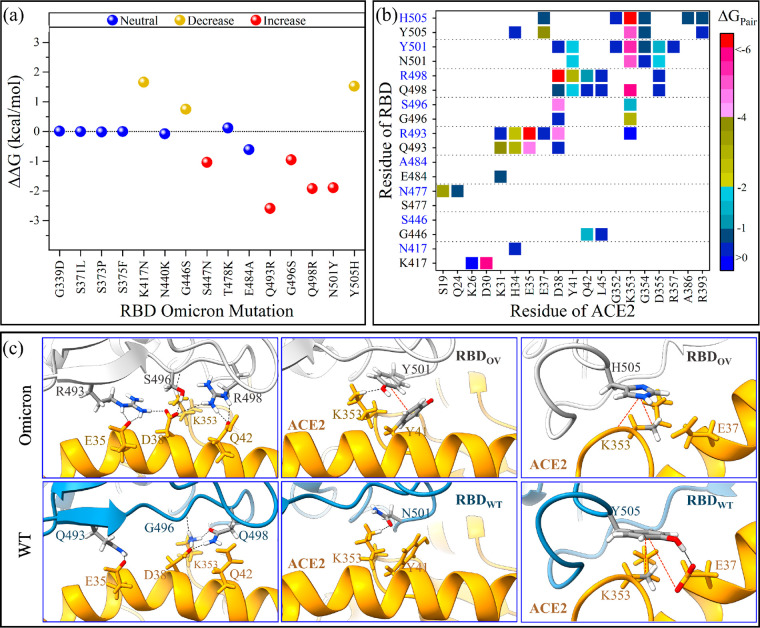
To gain a better understanding of the nature and impact of each mutation on the BFE in RBD-ACE2, in terms of per-residue fractions, decomposition schemes have been implemented and are shown in Figure S5 for all OV and WT single AAs. Figure 3a shows the per-residue BFE decomposition change for the 15 mutations between OV and WT (ΔΔG = ΔGOV – ΔGWT), while their AA–AA interaction pair maps are displayed in Figures 3b. These mutations are divided into three groups based on their influence on binding: neutral, decreased, and increased binding.
Figure 3.
BFE decompositions of RBD-ACE2 complex in OV and WT. (a) Change in per-residue BFE decomposition (ΔΔG = ΔGOV – ΔGWT). The 15 OV mutations are classified based on their ACE2 binding properties as neutral, increased, or decreased binding. (b) Pairwise BFE decomposition for only the mutated OV residues (blue characters on y-axis) that form pairs with ACE2, compared with their WT (black). (c) Details of the interactions for the five important mutations in OV (Q493R, G496S, Q493R, N501Y, and Y505H) and compared with their WT. The black dashed lines represent possible hydrogen bonds or salt-bridges, while the red dashed lines represent hydrophobic interactions.
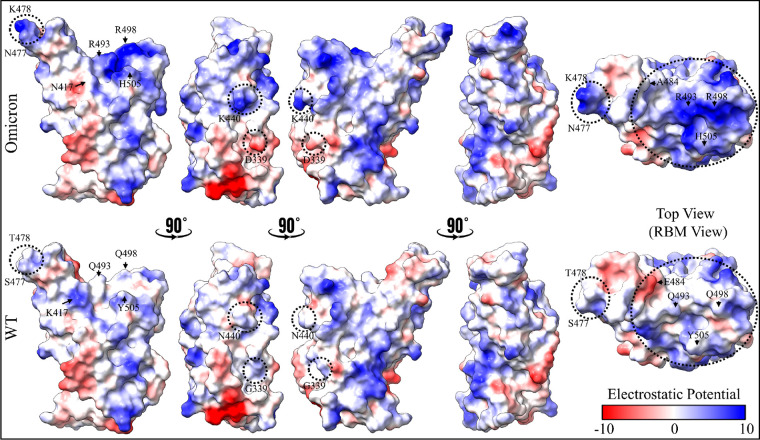
Although mutations outside of RBM and away from the interface, such as G339D, S371L, S373P, and S375F, do not affect the RBD–ACE2 binding, they may still give rise to other biological consequences. For instance, substituting neutral G339 with highly negatively charged D339 changes the surface charge distribution of RBD (Figure 4), which may impair the binding between RBD and antibody. Indeed, a recent study found that this mutation has a slightly higher escape fraction for Sotrovimab, a human neutralizing monoclonal antibody (mAb).41 Aside from that, changing G339 to D339 results in a longer side chain, which probably varies the local intramolecular interactions, particularly with the N343 glycosylation site.12 We did not include glycans in our simulation because earlier work suggested that the RBD of the S-protein had far less glycans than the S-protein itself and did not interact directly with ACE2.42 Mutating the polar residue (S) to the hydrophobic residues, L at 371, P at 373, and F at 375, forms a unique cluster that changes the biochemical properties of this RBD region in ways not previously observed in any other strains. This again allows OV to escape from the class 4 antibodies and some other antibodies from class 1, 2, and 3.11,12 Surprisingly, our MD simulations revealed that S371L, S373P, and S375F mutations are more flexible and induce a conformational change in RBD, suggesting that they have a higher chance to evade antibody recognition.
Figure 4.
Comparison of electrostatic potential surface of RBD in mutated Omicron (top) and WT (bottom). From left to right, the first four shapes are of the RBD front view, rotated sequentially by 90°, while the fifth one represents the RBD top view. Important residues that changed their electrostatic potential surfaces are labeled. Negative-potential residues are shown in red, near-neutral residues in white, and positive-potential residues in blue.
Our findings based on BFE decompositions show that the E484A, T478K, and N440K mutations would exhibit the same pattern of neutral binding (Figure 3). Interestingly, E484K mutation has also been observed in Beta and Gamma VOC and another variant of interest (VOI), and it has been identified as an immunodominant spike protein residue. Therefore, E484A in OV is expected to greatly reduce the susceptibility of many mAbs, which is fully consistent with the recent studies.11,12 In our MD simulations, we observed that the E484A mutation eliminates the weak E484:K31 in WT, but it reduces the destabilization that is stemming from possible electrostatic repulsion between E484 of WT and E35 of ACE2 when changing to A484. Therefore, this mutation has no impact on BFE, unlike the E484K mutation in Beta, which increases the interaction marginally.17 T478K has also been seen in the Delta variant and has no direct influence on the OV RBD-ACE2 interface network.43 Finally, the N440K has been linked to an increase in antibody neutralization resistance for some antibodies.11,44 Importantly, the exchange of amino acids at these sites (440, 478, and 484) contributes to the emergence of a different electrostatic surface of RBD, which may play some role in attractive electrostatic interactions with the negatively charged surface of ACE2 (Figure 4). In this context, ab initio quantum methodologies offer a more accurate description of the partial charge distributions for these charged residues.17,45−50
Similar to the Beta variant, the K417N mutation reduces the OV RBD-ACE2 binding because of the loss of the strong ionic pair with D30 on ACE2.17 While this result is consistent with previous observations,29,51 we note that the K417N mutation could affect also the way the RDB clamps the ACE2.19 Certain mAbs, such as Etesevimab and Casirivimab, have been shown to be affected by K417N.52 Similarly, the G446S and Y505H reduce the binding, but we suspect that this is because of the impact of other mutations like N501Y rather than the mutations themselves. In our early study, we observed that the N501Y in the Alpha and Beta VOC significantly enhances the interactions with ACE2, but it eliminates the hydrogen bond (HB) between the G446 and Q42 of ACE2 and reduces the strength of the Y505:E37 pair (Figure 3c).17 The same is true in OV, even though the residues at 446 and 505 site are now S and H, and as a result, their overall contributions to BFE in OV are smaller than in WT.
On the other hand, S477N, Q493R, G496S, Q498R, and N501Y mutations confer additional strength to the ACE2 binding, while N477 (OV) forms new HBs with S19 on ACE2. Interestingly, our findings indicate that the main source of increased binding is due to the formation of two new strong salt bridges between R493 and R498 of OV and E35 and D38 on ACE2 with pair interaction strengths of −11 and −6.5 kcal/mol, respectively, which was not observed in the WT (Figure 3b,c). According to our findings, the dominant mutations in the OV that enhance electrostatic interactions to ACE2 are Q493R and Q498R. This finding is in full agreement with experimental results.53,54 R493 (OV) can also form a salt bridge with D38 with pair interaction strength of −4.5 kcal/mol, but it also makes a weak unfavorable pair with K353 and loses the pair with K31 that exists in the WT. R498 (OV) retains the pairings with Y41 and Q42, but they are stronger than Q498 (WT). However, it loses the pair with K353 because of the N501Y mutation.17 Unlike G496 of WT, the aliphatic hydroxyl group of S496 (OV) also forms new HBs with D38 on ACE2 with pair interaction strength of −4.3 kcal/mol and maintains the HB with K353 (Figure 3b,c). Just like in Alpha and Beta VOC, Y501 forms more pairings than N501 of WT (Figure 3b,c).17 The number of pairs between Y501 of OV and ACE2 residues is the same as in Alpha and Beta, but their interaction strengths change, particularly the Y501:D38 pair (−0.35 of OV vs −4 kcal/mol of Alpha or Beta17).
In conclusion, comprehensive MD simulations have been performed to investigate the effects of OV RBD mutations on ACE2 human cell receptor binding. Our results shed light on the critical roles of the new OV mutations in the conformational changes of RBD (S371L, S373P, and S375F) and changes in its electrostatic potential surface (N440K, T478K, E484A, Q493R, Q498R, and Y505H). As a result, the overall electrostatic attraction between the positively charged OV RBD and the negatively charged ACE2 receptor is twice as strong as in the WT, leading to increased OV contagiousness as well as to the escape from neutralizing antibodies. Our analysis also shows that the loss of some interactions caused by K417N, G446S, and Y505H is completely compensated by the formation of new pairs from S477N, Q493R, G496S, Q498R, and N501Y, resulting in an overall stronger binding of OV’s RBD-ACE2.
Acknowledgments
This research used the resources of the National Energy Research Scientific Computing Center supported by DOE under Contract No. DE-AC03-76SF00098 and the Research Computing Support Services (RCSS) of the University of Missouri System. This project is funded by the National Science Foundation of USA: RAPID DMR/CMMT-2028803. R.P. acknowledges funding from the Key project #12034019 of the National Natural Science Foundation of China.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.2c00423.
Principal component analysis (Figure S1), time evaluation and the histogram distribution of BFE in the OV RBD-ACE2 complex (Figure S2), RMSF comparison between OV and WT (Figure S3), difference energetic components between OV and WT (Figure S4), and per-residue decomposition comparison between OV and WT (Figure S5) (PDF)
Transparent Peer Review report available (PDF)
Author Contributions
B.J. and W.-Y.C. conceived the project. B.J. performed the calculations and made most of the figures. B.J. and W.-Y.C. drafted the paper with input from P.A. and R.P. All authors participated in the discussion and interpretation of the results. All authors edited and proofread the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Harvey W. T.; Carabelli A. M.; Jackson B.; Gupta R. K.; Thomson E. C.; Harrison E. M.; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R.; Moyo S.; Amoako D. G.; Tegally H.; Scheepers C.; Althaus C. L.; et al. Rapid Epidemic Expansion of the SARS-CoV-2 Omicron Variant in Southern Africa. Nature 2022, 603, 679. 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S. S. A.; Karim Q. A. Omicron SARS-CoV-2 Variant: A New Chapter in the COVID-19 Pandemic. Lancet 2021, 398, 2126–2128. 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W.; Huo J.; Zhou D.; Zahradník J.; Supasa P.; Liu C.; et al. SARS-CoV-2 Omicron-B.1.1.529 Leads to Widespread Escape from Neutralizing Antibody Responses. Cell 2022, 185, 467–484. 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L.; Segovia-Chumbez B.; Jadi R.; Martinez D. R.; Raut R.; Markmann A. J.; Cornaby C.; Bartelt L.; Weiss S.; Park Y.; Edwards C. E.; Weimer E.; Scherer E. M.; Rouphael N.; Edupuganti S.; Weiskopf D.; Tse L. V.; Hou Y. J.; Margolis D.; Sette A.; Collins M. H.; Schmitz J.; Baric R. S.; de Silva A. M.; et al. The Receptor-Binding Domain of the Viral Spike Protein Is an Immunodominant and Highly Specific Target of Antibodies in SARS-CoV-2 Patients. Sci. Immunol. 2020, 5, eabc8413 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Yang C.; Xu X.; Xu W.; Liu S. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W.; He L.; Zhang X.; Pu J.; Voronin D.; Jiang S.; et al. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Wang W.; Chen Z.; Lu S.; Yang F.; Bi Z.; et al. A Vaccine Targeting the RBD of the S Protein of SARS-CoV-2 Induces Protective Immunity. Nature 2020, 586, 572–577. 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Hoffmann M.; Krüger N.; Schulz S.; Cossmann A.; Rocha C.; Kempf A.; et al. The Omicron Variant Is Highly Resistant against Antibody-Mediated Neutralization: Implications for Control of the COVID-19 Pandemic. Cell 2022, 185, 447–456. 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.; Mok B. W.-Y.; Chen L.-L.; Chan J. M.-C.; Tsang O. T.-Y.; Lam B. H.-S.; et al. Neutralization of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant by Sera From BNT162b2 or CoronaVac Vaccine Recipients. Clin. Infect. Dis. 2021, 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Wang J.; Jian F.; Xiao T.; Song W.; Yisimayi A.; et al. Omicron Escapes the Majority of Existing SARS-CoV-2 Neutralizing Antibodies. Nature 2022, 602, 657–663. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G.; Guo Y.; Liu L.; Zhang Z.; Liu L.; Luo Y. Structural Basis for Antibody Resistance to SARS-CoV-2 Omicron Variant. bioRxiv 2021, 2021.12.21.473620. 10.1101/2021.12.21.473620. [DOI] [Google Scholar]
- Cameroni E.; Bowen J. E.; Rosen L. E.; Saliba C.; Zepeda S. K.; Culap K.; et al. Broadly Neutralizing Antibodies Overcome SARS-CoV-2 Omicron Antigenic Shift. Nature 2022, 602, 664–670. 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C.; Evans J. P.; Qu P.; Faraone J.; Zheng Y.-M.; Carlin C.; et al. Neutralization and Stability of SARS-CoV-2 Omicron Variant. bioRxiv 2021, 2021.12.16.472934. 10.1101/2021.12.16.472934. [DOI] [Google Scholar]
- Planas D.; Saunders N.; Maes P.; Guivel-Benhassine F.; Planchais C.; Buchrieser J.; et al. Considerable Escape of SARS-CoV-2 Omicron to Antibody Neutralization. Nature 2022, 602, 671–675. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Tao K.; Tzou P. L.; Nouhin J.; Gupta R. K.; de Oliveira T.; Kosakovsky Pond S. L.; et al. The Biological and Clinical Significance of Emerging SARS-CoV-2 Variants. Nat. Rev. Genet. 2021, 22, 757–773. 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad B.; Adhikari P.; Podgornik R.; Ching W. Y. Key Interacting Residues between RBD of SARS-CoV-2 and ACE2 Receptor: Combination of Molecular Dynamics Simulation and Density Functional Calculation. J. Chem. Inf. Model. 2021, 61, 4425–4441. 10.1021/acs.jcim.1c00560. [DOI] [PubMed] [Google Scholar]
- Spinello A.; Saltalamacchia A.; Magistrato A. Is the Rigidity of SARS-CoV-2 Spike Receptor-Binding Motif the Hallmark for Its Enhanced Infectivity? Insights from All-Atom Simulations. J. Phys. Chem. Lett. 2020, 11, 4785–4790. 10.1021/acs.jpclett.0c01148. [DOI] [PubMed] [Google Scholar]
- Spinello A.; Saltalamacchia A.; Borišek J.; Magistrato A. Allosteric Cross-Talk among Spike’s Receptor-Binding Domain Mutations of the SARS-CoV-2 South African Variant Triggers an Effective Hijacking of Human Cell Receptor. J. Phys. Chem. Lett. 2021, 12, 5987–5993. 10.1021/acs.jpclett.1c01415. [DOI] [PubMed] [Google Scholar]
- Casalino L.; Gaieb Z.; Goldsmith J. A.; Hjorth C. K.; Dommer A. C.; Harbison A. M.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2020, 6, 1722–1734. 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Liu Y.; Lei Z.; Dicker J.; Cao Y.; Zhang X. F.; et al. Differential Interactions between Human ACE2 and Spike RBD of SARS-CoV-2 Variants of Concern. J. Chem. Theory Comput. 2021, 17, 7972–7979. 10.1021/acs.jctc.1c00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler M.; Ray A.; Moreira R. A.; Juniku B.; Poma A. B.; Alsteens D. Molecular Insights into Receptor Binding Energetics and Neutralization of SARS-CoV-2 Variants. Nat. Commun. 2021, 12, 6977. 10.1038/s41467-021-27325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin M.; Sorour M. K.; Kasry A. Comparing the Binding Interactions in the Receptor Binding Domains of SARS-CoV-2 and SARS-CoV. J. Phys. Chem. Lett. 2020, 11, 4897–4900. 10.1021/acs.jpclett.0c01064. [DOI] [PubMed] [Google Scholar]
- Goher S. S.; Ali F.; Amin M. The Delta Variant Mutations in the Receptor Binding Domain of SARS-CoV-2 Show Enhanced Electrostatic Interactions with the ACE2. Med. Drug Discovery 2022, 13, 100114. 10.1016/j.medidd.2021.100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali F.; Kasry A.; Amin M. The New SARS-CoV-2 Strain Shows a Stronger Binding Affinity to ACE2 Due to N501Y Mutant. Med. Drug Discovery 2021, 10, 100086. 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Shapovalov M. V.; Dunbrack R. L. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure 2011, 19, 844–858. 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; et al. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Cai Y.; Zhang J.; Xiao T.; Lavine C. L.; Rawson S.; Peng H.; Zhu H.; Anand K.; Tong P.; Gautam A.; Lu S.; Sterling S. M.; Walsh R. M.; Rits-Volloch S.; Lu J.; Wesemann D. R.; Yang W.; Seaman M. S.; Chen B.; et al. Structural Basis for Enhanced Infectivity and Immune Evasion of SARS-CoV-2 Variants. Science 2021, 373, 642–648. 10.1126/science.abi9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Ginn H. M.; Dejnirattisai W.; Supasa P.; Wang B.; Tuekprakhon A.; et al. Reduced Neutralization of SARS-CoV-2 B.1.617 by Vaccine and Convalescent Serum. Cell 2021, 184, 4220–4236. 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman D. A.; Case D. A.; Caldwell J. W.; Ross W. S.; Cheatham T. E.; DeBolt S.; et al. AMBER, a Package of Computer Programs for Applying Molecular Mechanics, Normal Mode Analysis, Molecular Dynamics and Free Energy Calculations to Simulate the Structural and Energetic Properties of Molecules. Comput. Phys. Commun. 1995, 91, 1–41. 10.1016/0010-4655(95)00041-D. [DOI] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Maier J. A.; Martinez C.; Kasavajhala K.; Wickstrom L.; Hauser K. E.; Simmerling C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.; Sun H.; Wang J.; Wang Z.; Liu H.; Zhang J. Z. H.; et al. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- Miller B. R.; McGee T. D.; Swails J. M.; Homeyer N.; Gohlke H.; Roitberg A. E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Jawad B.; Poudel L.; Podgornik R.; Ching W. Y. Thermodynamic Dissection of the Intercalation Binding Process of Doxorubicin to DsDNA with Implications of Ionic and Solvent Effects. J. Phys. Chem. B 2020, 124, 7803–7818. 10.1021/acs.jpcb.0c05840. [DOI] [PubMed] [Google Scholar]
- Jawad B.; Poudel L.; Podgornik R.; Steinmetz N. F.; Ching W. Y. Molecular Mechanism and Binding Free Energy of Doxorubicin Intercalation in DNA. Phys. Chem. Chem. Phys. 2019, 21, 3877–3893. 10.1039/C8CP06776G. [DOI] [PubMed] [Google Scholar]
- Yin W.; Xu Y.; Xu P.; Cao X.; Wu C.; Gu C.; et al. Structures of the Omicron Spike Trimer with ACE2 and an Anti-Omicron Antibody. Science 2022, 375, 1048–1053. 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese L.; Zaccaria M.; Farzan M.; Johnson W.; Momeni B. Investigating the Mutational Landscape of the SARS-CoV-2 Omicron Variant via Ab Initio Quantum Mechanical Modeling. bioRxiv 2021, 2021.12.01.470748. 10.1101/2021.12.01.470748. [DOI] [Google Scholar]
- Lan J.; He X.; Ren Y.; Wang Z.; Zhou H.; Fan S.; et al. Structural and Computational Insights into the SARS-CoV-2 Omicron RBD-ACE2 Interaction. bioRxiv 2022, 2022.01.03.474855. 10.1101/2022.01.03.474855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart A. L.; Havenar-Daughton C.; Lempp F. A.; Ma D.; Schmid M. A.; Agostini M. L.; et al. The Dual Function Monoclonal Antibodies VIR-7831 and VIR-7832 Demonstrate Potent in Vitro and in Vivo Activity against SARS-CoV-2. bioRxiv 2021, 2021.03.09.434607. 10.1101/2021.03.09.434607. [DOI] [Google Scholar]
- Barros E. P.; Casalino L.; Gaieb Z.; Dommer A. C.; Wang Y.; Fallon L.; et al. The Flexibility of ACE2 in the Context of SARS-CoV-2 Infection. Biophys. J. 2021, 120, 1072–1084. 10.1016/j.bpj.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M.; Walls A. C.; Sprouse K. R.; Bowen J. E.; Rosen L. E.; Ha V.; Dang A. D. M.; et al. Molecular Basis of Immune Evasion by the Delta and Kappa SARS-CoV-2 Variants. Science 2021, 374, 1621–1626. 10.1126/science.abl8506. [DOI] [PubMed] [Google Scholar]
- Rani P. R.; Imran M.; Lakshmi J. V.; Jolly B.; Jain A.; Surekha A.; et al. Symptomatic Reinfection of SARS-CoV-2 with Spike Protein Variant N440K Associated with Immune Escape. J. Med. Virol. 2021, 93, 4163–4165. 10.1002/jmv.26997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari P.; Podgornik R.; Jawad B.; Ching W. Y. First-Principles Simulation of Dielectric Function in Biomolecules. Materials 2021, 14, 5774. 10.3390/ma14195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching W. Y.; Adhikari P.; Jawad B.; Podgornik R. Ultra-Large-Scale Ab Initio Quantum Chemical Computation of Bio-Molecular Systems: The Case of Spike Protein of SARS-CoV-2 Virus. Comput. Struct. Biotechnol. J. 2021, 19, 1288–1301. 10.1016/j.csbj.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari P.; Jawad B.; Rao P.; Podgornik R.; Ching W.-Y. Delta Variant with P681R Critical Mutation Revealed by Ultra-Large Atomic-Scale Ab Initio Simulation: Implications for the Fundamentals of Biomolecular Interactions. Viruses 2022, 14, 465. 10.3390/v14030465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari P.; Li N.; Shin M.; Steinmetz N. F.; Twarock R.; Podgornik R.; et al. Intra- And Intermolecular Atomic-Scale Interactions in the Receptor Binding Domain of SARS-CoV-2 Spike Protein: Implication for ACE2 Receptor Binding. Phys. Chem. Chem. Phys. 2020, 22, 18272–18283. 10.1039/D0CP03145C. [DOI] [PubMed] [Google Scholar]
- Baral K.; Adhikari P.; Jawad B.; Podgornik R.; Ching W.-Y. Solvent Effect on the Structure and Properties of RGD Peptide (1FUV) at Body Temperature (310 K) Using Ab Initio Molecular Dynamics. Polymers 2021, 13, 3434. 10.3390/polym13193434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad B.; Adhikari P.; Cheng K.; Podgornik R.; Ching W.-Y. Computational Design of Miniproteins as SARS-CoV-2 Therapeutic Inhibitors. Int. J. Mol. Sci. 2022, 23, 838. 10.3390/ijms23020838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan B.; Huynh T. Insights into SARS-CoV-2’s Mutations for Evading Human Antibodies: Sacrifice and Survival. J. Med. Chem. 2022, 65, 2820–2826. 10.1021/acs.jmedchem.1c00311. [DOI] [PubMed] [Google Scholar]
- Wang P.; Nair M. S.; Liu L.; Iketani S.; Luo Y.; Guo Y.; et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Mannar D.; Saville J. W.; Zhu X.; Srivastava S. S.; Berezuk A. M.; Tuttle K. S.; Marquez A. C.; Sekirov I.; Subramaniam S.; et al. SARS-CoV-2 Omicron Variant: Antibody Evasion and Cryo-EM Structure of Spike Protein–ACE2 Complex. Science 2022, 375, 760–764. 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P.; Li L.; Liu S.; Wang Q.; Zhang D.; Xu Z.; et al. Receptor Binding and Complex Structures of Human ACE2 to Spike RBD from Omicron and Delta SARS-CoV-2. Cell 2022, 185, 630–640. 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.