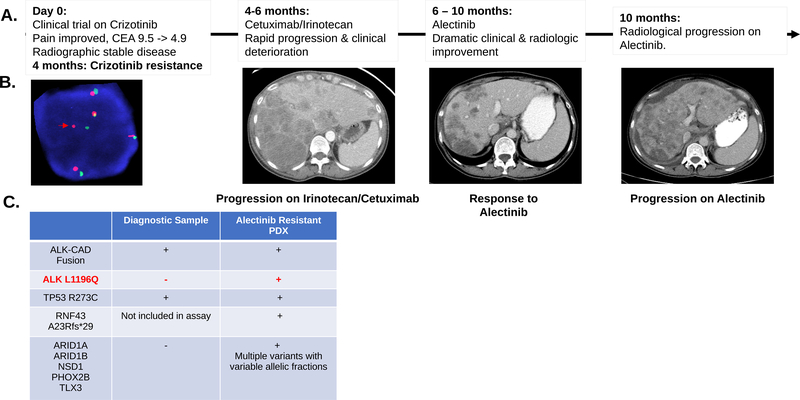
Figure 3.
A. Clinical and radiographic correlates of patient treatment history and response. The patient experienced a dramatic albeit short-lived response on alectinib. B. Break-apart FISH probes demonstrated the continued presence of ALK fusion in the alectinib-resistant tumor-derived xenograft. Red and green signals hybridize to the 5’ and 3’ ends of the ALK gene. In the case of an intact ALK gene the two signals map close together, often appearing as a yellow, or fused, signal. Rearranged alleles are scored when at least a two-signal diameter spacing is observed between the red and green ALK signals (Red arrow). In addition to ALK rearrangement the FISH assay shows evidence of ALK gene polysomy as evidenced by 5 copies of ALK gene in the representative image. C. Comparison of SNVs detected in diagnostic and alectinib-resistant samples for this index case. Genetic testing showed a shared TP53 mutation and the continued detection of ALK-CAD fusion. An acquired ALK L1196Q gatekeeper mutation was detected in the alectinib-resistant sample along with several other mutations at low allelic frequency (Suppl. Table 3). RNF43 frameshift (fs) mutation was detected in the alectinib-resistant sample. This gene was not part of the earlier version of the assay performed on the diagnostic sample.