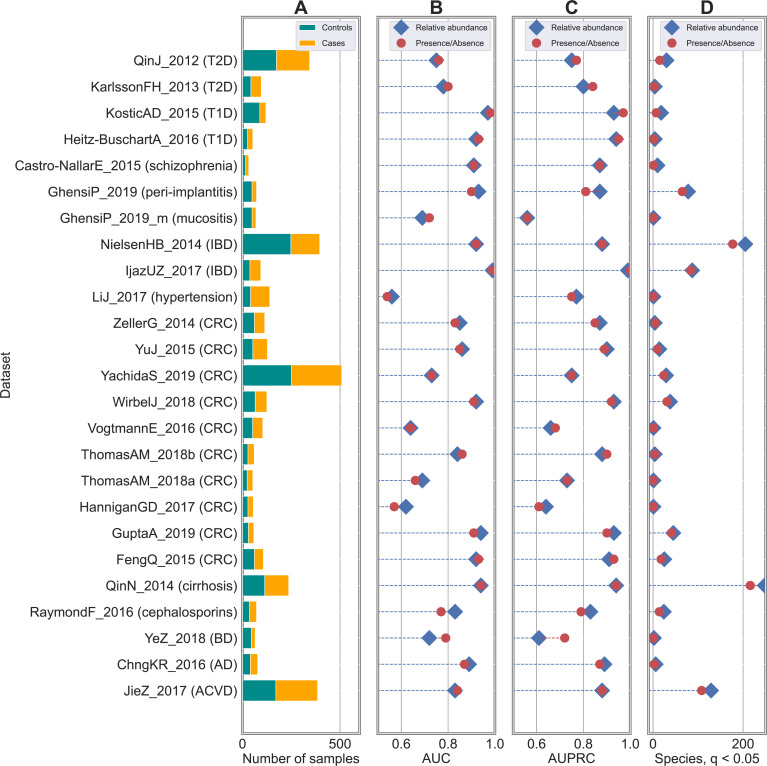
Fig 1. Classification accuracies are robust to degradation from species-level relative abundance to presence/absence profiles in shotgun datasets.
Results obtained on 25 case-control studies for host phenotype classification from human microbiomes. (A) Number of case and control samples across the different studies. (B) AUC and (C) AUPRC scores using RF as back-end classifiers on species-level taxonomic profiles. Comparison between relative abundance (in blue) and presence/absence (in red) profiles highlighted negligible differences and no statistical differences in none of the studies (see S1 Fig for AUC scores and S2 Table for p-values). Metrics of comparison in terms of AUC, AUPRC, precision, recall, and F1 are summarized in S2 Fig and S2 Table is represented a comparison between AUC and AUPCR scores. (D) Number of statistically significant taxa from relative abundance (in blue) and presence/absence (in red) profiles.