Figure 3. Determinants of tumor sensitivity to GLS inhibitors.
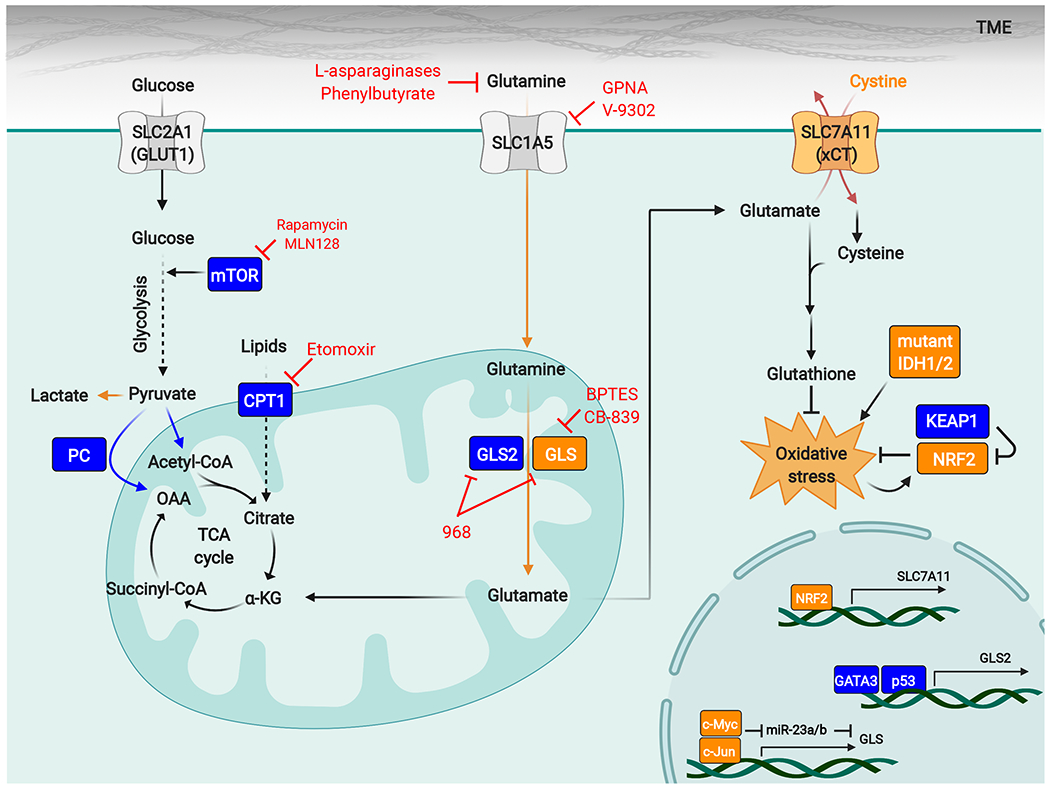
Diverse factors affect the sensitivity of tumors to GLS inhibitors, and specific oncogenotypes and metabolic stresses (shown in orange) can lock cancer cells into a GLS-dependent state. High activity of the NRF2-antioxidant response pathway has emerged as a conserved biomarker for tumor GLS dependence and can occur through genetic activation of the pathway (for example by loss-of-function mutations in KEAP1) or through factors that impose sustained oxidative stress, such as IDH1/2 mutations that lead to neomorphic 2-hydroxyglutarate-producing enzymes. Expression of GLS is regulated by c-Myc and c-Jun, and high levels of these transcription factors also favor cellular dependence on GLS. The nutrient environment also influences sensitivity to GLS inhibitors, and excessive levels of extracellular cystine can drive glutamine addiction and GLS dependence by forcing glutamate efflux through the cystine/glutamate antiporter xCT/SLC7A11. Other factors promote resistance to GLS inhibitors (shown in blue). These factors include compensatory metabolic pathways such as pyruvate carboxylation, fatty acid oxidation, and glutamine hydrolysis catalyzed by GLS2. mTORC1 can enhance metabolic flexibility by regulating a shift to glucose metabolism, and mTORC1 inhibitors synergize with GLS inhibitors.
Abbreviations: α-KG, α-ketoglutarate; CPT1, carnitine palmitoyltransferase I; GCL, glutamate-cysteine ligase; GLS, glutaminase; GPNA, L-γ-glutamyl-p-nitroanilide; IDH, isocitrate dehydrogenase; KEAP1, Kelch-like ECH-associated protein 1; mTOR, mechanistic target of rapamycin; NRF2, nuclear factor erythroid 2-related factor 2; OAA, oxaloacetate; PC, pyruvate carboxylase; ROS, reactive oxygen species; TME, tumor microenvironment.
