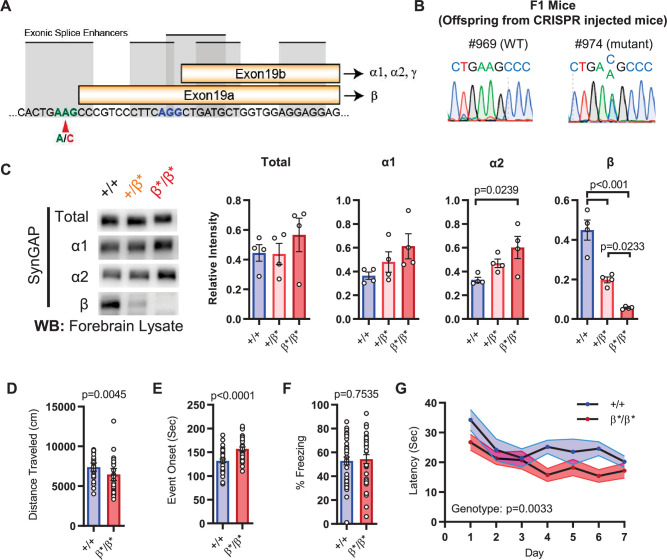
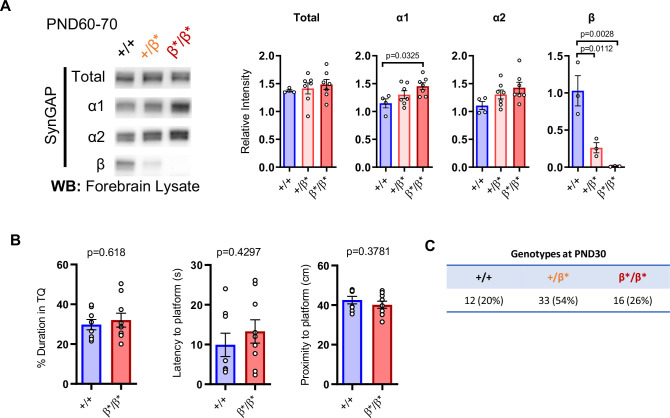
Figure 3. Design and characterization of Syngap1β* knock-in mice.
(A) Alternative use of exon19 in distinct splicing events. Exon19 can be spliced into 2 frames shifted by 13 bp. Use of early splice acceptor (green) results in a frameshift and expresses β isoform. Use of the late splice acceptor (blue) allows expression of all other SynGAP C-terminal variants. To specifically disrupt SynGAP-β, a point mutation (A to C) was introduced to the early splice acceptor (indicated with red arrow). (B) Sequence trace of Syngap1β*/+ mice obtained via crossing F0 founders to wild-type mice. Mutation site exhibits equal levels of A and C signal in sequence trace indicating heterozygosity. (C) Representative western blots showing expression levels of total SynGAP and individual isoforms at PND7. Relative intensity of bands normalized to total protein signal. ANOVA with Tukey’s multiple comparisons test. Total: F(2, 9) = 0.7427, p = 0.5029. α1: F(2, 9) = 2.391, p = 0.147. α2: F(2, 9) = 5.333, p = 0.0297. β: F(2, 9) = 42.53, p < 000.1(D) Quantification of total distance traveled in OFT. +/+ (n = 36), β/β (n = 32); Mann-Whitney U = 346, p = 0.0045. (E) Seizure threshold was measured as the time taken to reach three separate events of 1st clonus (event onset) during the procedure. Unpaired t-test t(66)=4.237. (F) Percent freezing in remote contextual fear memory paradigm. % Freezing: t(66)=0.3153. (G) Plots demonstrating latency to find platform across days in Morris Water Maze training session. Statistical significance was determined by using linear mixed model for repeated measures. Genotype: F(1, 15) = 12.22, p = 0.0033.