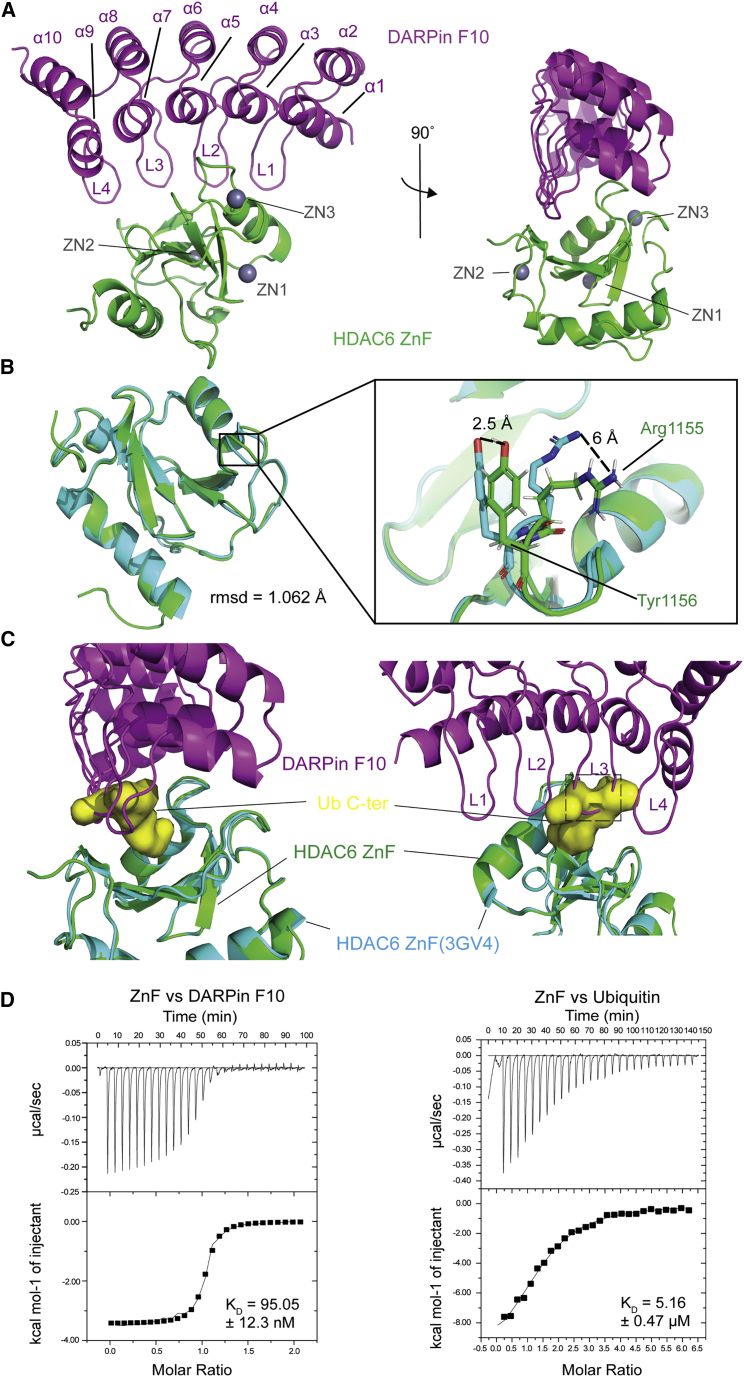
Figure 2.
2.55-Å crystal structure shows the detail of DARPin F10 inhibition on Ub recruitment
(A) F10 bound to HDAC6 ZnF. F10, ZnF (aa 1,108–1,215), and Zn2+ ions are colored purple, green, and gray. F10 α helices (α1 to α10) and β turns (L1 to L4) are numbered starting from the N-terminus (PDB: 7ATT).
(B) ZnF conformation alignment in two binding forms. Green shows the ZnF structure bound to F10, and cyan shows the ZnF bound to Ub (PDB: 3GV4). Right: zoomed-in view of the Ub binding “gatekeeper” amino acids, Arg1155 and Tyr1156. The shifting of side chains is indicated by dashed lines with the distance labeled.
(C) Zoomed-in views of the ZnF-Ub-binding pocket. The ZnF-F10 structure is superimposed onto the ZnF-Ub C-terminal tail structure (PDB: 3GV4). The Ub C-terminal tail is shown in surface representation (yellow): F10 L3 backbone clashes with the Ub tail. The region from His101 to Gly103 is indicated by the dashed line rectangle (right).
(D) Affinities of ZnF for F10 or free Ub by ITC assays. ZnF-F10: ΔS = 20.6 cal/mol/deg, N = 1.01. ZnF-Ub: ΔS = −12.8 cal/mol/deg, N = 1.46.