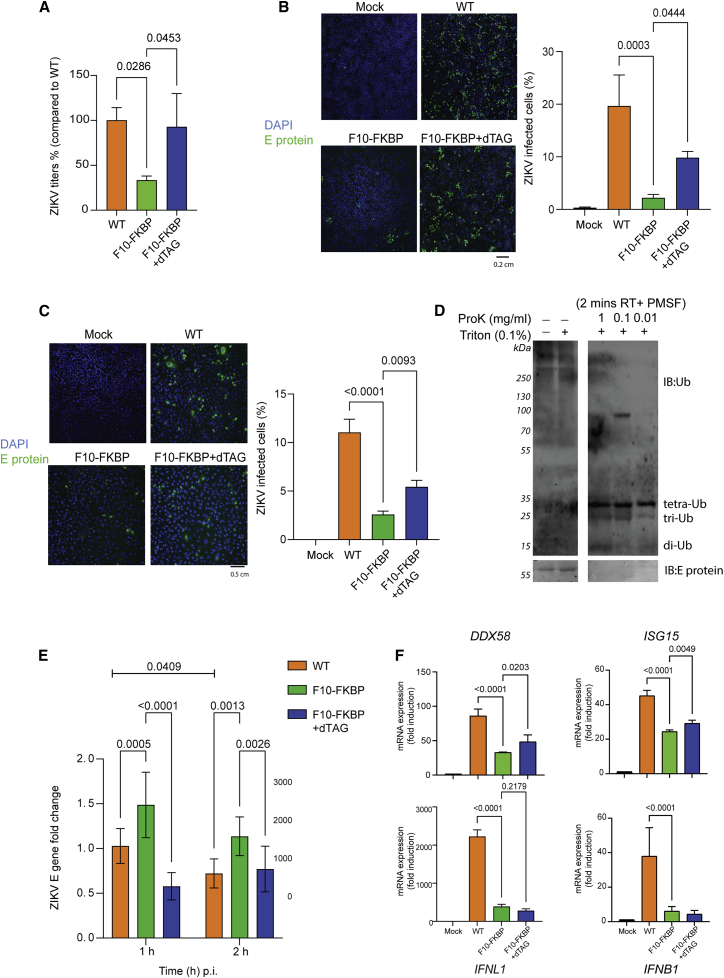
Figure 4.
ZIKAV replication is inhibited by DARPin F10
(A) Reduction of ZIKV titer in F10-expressing cells. The indicated cell lines (−/+ dTAG pre-treatment) were infected with ZIKV at an MOI of 0.1 50% tissue culture infective dose (TCID50)/cell, and culture supernatant was analyzed at 72 h. Viral titers were determined by a TCID50 assay; the baseline titer obtained with A549 WT cells was set to 100%. Statistical analysis was done with one-way ANOVA test; p values refer to the significant (< 0.05) differences between samples (n = 3). Data are represented as mean ± SD.
(B) Reduced ZIKV infection in F10-FKBP cells. Left panels: microscopy visualization of ZIKV E protein expression. After ZIKV titer quantification in (A), the cells at 72 hpi were stained by DAPI (blue) and for ZIKV E protein (green). 10× objective; scale bar, 0.2 cm. Mock WT refers to non-infected A549 cells; all other samples were infected. Right graph: quantification of the ZIKV E protein-positive cells based on independent experiments (n = 3). Statistical analysis as in (A).
(C) ZIKV infection is halted by F10 at early time point. Analysis and quantification as in (B) but at 16 hpi. 20× objective.
(D) Ub is present in ZIKV particles. Purified ZIKV particles (African strain) were incubated with Proteinase K (ProK) or Triton X-100. Samples were analyzed by immunoblotting with antibodies detecting Ub and ZIKV E protein.
(E) Elevated ZIKV RNA early after infection in cells expressing F10. The indicated cells (−/+ dTAG pre-treatment) were infected with ZIKV at high MOI (= 10) and RNA was analyzed at 1 and 2 hpi. ZIKV E genes RNA level is shown as fold change after normalization to 18S and Actin RNA controls; WT A549 (1 h) was used as baseline for relative comparison with 2−ΔΔct method (n = 3). Statistical analysis was done with two-way ANOVA; p values (< 0.05) are shown. Data are represented as mean ± SD.
(F) No antiviral genes upregulation in ZIKV-infected F10-FKBP cells. The indicated cell lines (−/+ dTAG pre-treatment) were infected with ZIKV (MOI = 1), and total RNA was extracted at 24 hpi. Expression of interferon-related genes, DDX58 (RIG-I), ISG15 (ISG15), IFNL1 (IL-29 or IFN-λ), and IFNB1 (IFN-β1) was analyzed by qRT-PCR (n = 3). Actin and GAPDH served as double control. Mock (uninfected WT A549 cells) was used as baseline for relative comparison with 2−ΔΔct method. Statistical analysis as in (A); difference to Mock not shown.