Figure 2. Nutritional conditions affect the chromatin landscape.
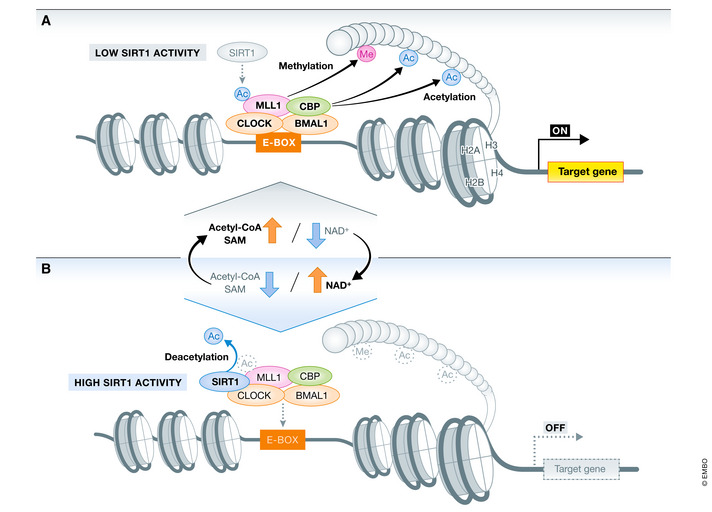
Most chromatin‐modifying enzymes use intermediary metabolites as cofactors or substrates, and their activity is regulated by the availability of these metabolites. Acetyl‐CoA, SAM, and NAD⁺ are substrates for histone acetylation, methylation, and deacetylation, respectively. Acetyl‐CoA is necessary for protein acetylation. SAM is an important methyl‐group donor metabolite in DNA and histone methylation. SIRT1 deacetylates histone lysine residues using NAD+ as a co‐substrate. (A) MLL1 physically interacts with CLOCK and contributes to CLOCK:BMAL1 recruitment to the E‐box regions. Together, MLL1 and CLOCK induce histone modifications (H3K4me4, H3K9Ac, and H3K14Ac) and enhance target gene expression. (B) Oscillating levels of NAD⁺ control SIRT1 deacetylase activity (histone and non‐histone proteins). Under high cellular NAD⁺ conditions, SIRT1 deacetylates MLL1, an event that reduces MLL1 enzymatic activity and leads to decreased H3K4me3 and H3K9/14ac levels. NAD⁺ thus controls MLL1 acetylation level and CLOCK:BMAL1 dependent transcription. BMAL1, brain and muscle ARNT‐like 1; CLOCK, circadian locomotor output cycles protein caput; MLL1, mixed‐lineage leukemia 1; NAD⁺, oxidized nicotinamide adenine dinucleotide; SAM, s‐adenosyl methionine; SIRT1, sirtuin 1.
