Fig. 3.
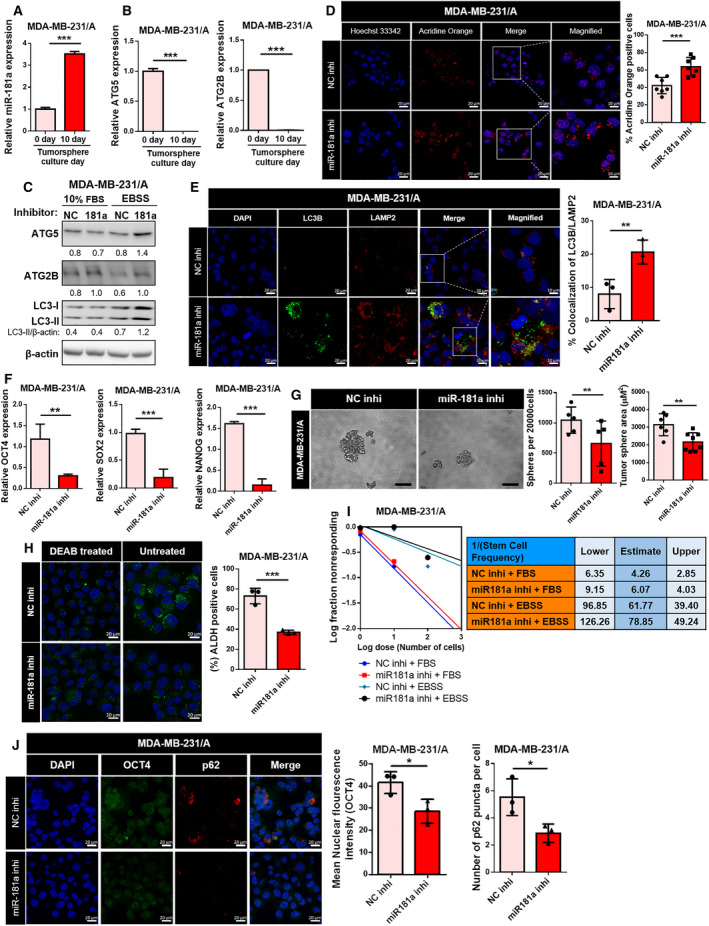
Inhibition of miR‐181a led to changes in autophagy and cancer stemness. (A) Expression levels of miR‐181a in MDA‐MB‐231/A tumorspheres were assessed using Taqman qRT‐PCR on days 0–10 of tumorsphere culture. Data were presented as mean ± SD of three independent experiments. Statistical analyses were performed with one‐tailed Student’s t‐test (***P < 0.001). (B) ATG5 and ATG2B mRNA levels in MDA‐MB‐231/A tumorspheres. Data were presented as mean ± SD of three independent experiments. Statistical analyses were performed with one‐tailed Student’s t‐test (***P < 0.001). (C) Expression levels of ATG5, ATG2B, and LC3 proteins in cells transfected with miR‐181a inhibitor. NC, negative control. (D) Confocal microscopy of acridine orange staining in MDA‐MB‐231/A tumorspheres (left, magnification: ×40, zoom: 2.0). The scale bar for magnified images equals 10 μm. Nuclei were counterstained with Hoechst 33342. Scale bar, 20 μm. The percentage of MDA‐MB‐231/A cells containing acridine orange puncta is shown (right). The number of nuclei was used to normalize the values. Data were presented as mean ± SD. Statistical analyses were performed with one‐tailed Student’s t‐test (***P < 0.001). (E) Confocal microscopy of LC3B and LAMP2 staining in tumorspheres (left, magnification: ×40, zoom: 2.0). The scale bar for magnified images equals 10 μm. Nuclei were counterstained with DAPI. Scale bar, 20 μm. Colocalization of LC3B and LAMP2 puncta was quantified, and normalized with the number of all measured puncta (right). Data were presented as mean ± SD. Statistical analyses were performed with one‐tailed Student’s t‐test (**P < 0.01). (F) The mRNA levels of OCT4, SOX2, and NANOG in MDA‐MB‐231/A cells after transfection with miR‐181a inhibitors were measured using qRT‐PCR. Data were presented as mean ± SD of four independent experiments. Statistical analyses were performed with one‐tailed Student’s t‐test (**P < 0.01, ***P < 0.001). (G) Representative images of MDA‐MB‐231/A tumorspheres transfected with miR‐181a inhibitors (left, magnification: ×20). Scale bar, 100 μm. The number and sizes of tumorspheres were quantified (right). Data were presented as mean ± SD. Statistical analyses were performed with one‐tailed Student’s t‐test (**P < 0.01). (H) MDA‐MB‐231/A cells transfected with miR‐181a inhibitor were stained with Aldefluor reagent (left). Cells treated additionally with DEAB (Aldefluor inhibitor) were used as a negative control. Nuclei were counterstained with Hoechst 33342 (Hoechst 33342; blue). Scale bar, 20 μm. The percentage of DEAB‐untreated MDA‐MB‐231/A cells containing ALDH (Aldefluor; green) puncta is shown (right). The number of nuclei was used to normalize the values. Data were presented as mean ± SD. Statistical analyses were performed with one‐tailed Student’s t‐test (***P < 0.001). (I) Limiting dilution assay performed on MDA‐MB‐231/A cells transfected with miR‐181a inhibitors. Stem cell frequency was calculated using extreme limiting dilution assay analysis. (J) Immunocytochemistry staining for detection of p62 and OCT4 in MDA‐MB‐231/A cells transfected with miR‐181a inhibitor (left, magnification: ×40). Nuclei were counterstained with DAPI. Scale bar, 20 μm. Mean nuclear intensity of OCT4 fluorescence is shown (right). The number of p62 puncta was normalized with the number of DAPI‐stained nuclei (right). Data were presented as mean ± SD. Statistical analyses were performed with one‐tailed Student’s t‐test (*P < 0.05).
