Abstract
Due to their involvement in numerous biochemical pathways, neuropeptides have been the focus of many recent research studies. Unfortunately, classic analytical methods, such as Western blots and enzyme-linked immunosorbent assays, are extremely limited in terms of global investigations, leading researchers to search for more advanced techniques capable of probing the entire neuropeptidome of an organism. With recent technological advances, mass spectrometry (MS) has provided methodology to gain global knowledge of a neuropeptidome on a spatial, temporal, and quantitative level. This review will cover key considerations for the analysis of neuropeptides by MS, including sample preparation strategies, instrumental advances for identification, structural characterization, and imaging; insightful functional studies; and newly developed absolute and relative quantitation strategies. While many discoveries have been made with MS, the methodology is still in its infancy. Many of the current challenges and areas that need development will also be highlighted in this review.
Keywords: Mass spectrometry, imaging, neuropeptides, microdialysis, post-translational modifications, quantitation
Introduction
The nervous system is one of the most highly regulated parts of the human body, and signaling molecules are well known for their roles in behavior, controlling bodily homeostasis, and processing incoming information (Herlenius & Lagercrantz, 2004; Hokfelt et al., 2000; Li & Sweedler, 2008; Xie, Romanova & Sweedler, 2011). Any perturbation of this system can have detrimental effects on an organism, leading to temporary or long-term biochemical changes. Neuropeptides, one of the largest classes of neuronal signaling molecules, are well known for playing prominent roles in the nervous system (Herlenius & Lagercrantz, 2004; Hokfelt et al., 2000; Li & Sweedler, 2008; Xie, Romanova & Sweedler, 2011). However, the comprehensive analyses of the neuropeptidome, the entire range of neuropeptides able to be expressed, remain to be challenging due to global diversity of their size, sequence, and function.
The diversity of neuropeptides can be first observed at the biological synthesis level. This review only focuses on the typical neuropeptide biosynthesis pathway and not on alternative ways of endogenous peptide production. A typical neuropeptide biosynthesis starts with the translation of a prepropeptide RNA chains. A prepropeptide may contains several neuropeptide copies, which are revealed after multiple processing steps. Initially, a propeptide is produced from the prepropeptide via proteolytic cleavages, splicing events, or introduction of post-translational modifications (PTMs) (Li & Sweedler, 2008). The result is a propeptide which is packaged into vesicles where they are stored prior to release. A strong stimulation, such as high frequency firing, elicits site-specific enzymes to produce the final, biologically active peptides that are released from the neuron. Mature neuropeptides released in the extracellular space 'travel' through the body to reach (distant) organs/tissues/cells which contain receptors where they bind. The latter are sometimes referred to as neuropeptide targets. The final neuropeptides generally range in length from 3 to 70 amino acids long (Buchberger, Yu & Li, 2015). The signaling targets can be within the same neuron produced, within the same organ, or in an entirely different tissue. In addition, neuropeptide anabolism, catabolism, and thus function may even vary depending on the destination of the signaling target (von Bohlen und Halbach, 2005). To further increase the chemical diversity, neuropeptides can have isoforms that may only vary by one residue but have widely different functions within the body. All these factors lead to a high, natural complexity that is difficult to characterize even with complete genetic coverage.
The development of sophisticated analytical tools or simplified networks are required for deep neuropeptidomic analysis. To decrease the complexities of neuropeptide analysis, many researchers have adopted different, similar animal models, such as crustaceans or mice, to characterize neuropeptidomic changes (Che et al., 2005; Chen et al., 2014; OuYang, Liang & Li, 2015; Yin et al., 2011; Zhang et al., 2015). Due to homology between neuropeptides from different species, many of the results and insights obtained from these simpler systems can be readily transferred to more complex organisms, such as humans (Bruzzone et al., 2006; Schmerberg & Li, 2013; Yew et al., 2005; Yu et al., 2014). As the full complement of neuropeptides has yet to be fully discovered, even with the aid of these model organisms, it is important to develop and implement more advanced technology.
To fully characterize neuropeptides, we require methodology that is selective, sensitive, and swift, all while being cost-effective and capable of providing dynamic temporal and spatial information. In the past, researchers have focused on the use of antibody-based, electrochemical, bioluminescent, or other biological assays to characterize neuropeptides (Li & Sweedler, 2008). For example, radioimmunoassays (RIAs) were very popular at one time due to being highly sensitive and selective (Li & Sweedler, 2008), even to familial isoforms (Jarecki et al., 2013), but their high cost and inability to simultaneously study multiple analytes, spatially and quantitatively, limits their global use. Unlike these classical methods, mass spectrometry (MS) has begun to meet all the necessary requirements for scientists to fully study neuropeptides. In general, MS measures the mass-to-charge ratio (m/z) of an analyte of interest. These instruments are capable of analyzing neuropeptides down to low attomole ranges while providing mass accuracy down to a few ppm and resolution to differentiate between not only different neuropeptides but also familial isoforms (Andren, Emmett & Caprioli, 1994; Dowell, Heyden & Li, 2006; Hui et al., 2012). While the development of high-resolution, accurate mass (HRAM) instrumentation allows for identification at the single stage MS (MS1) level, masses can also be selected for tandem MS (MS/MS). Peptide precursor ions are fragmented, producing characteristic fragments. As such, both known and novel analytes can be characterized and/or confidently identified. In conjunction with online or offline separations, MS is claimed to be capable of analyzing “entire proteomes” in a short amount of time (Hebert et al., 2014), making it an excellent tool to study the full complement of neuropeptides in a system (Castro et al., 2014; Hui et al., 2013; Predel et al., 2018; Predel et al., 2010; Xie, Romanova & Sweedler, 2011). Furthermore, the development of MS imaging has allowed to obtain highly accurate spatial information of several hundred analytes in one experiment. In addition, several strategies have been also developed (label-free and label-based) to quantitatively study neuropeptide changes, such as due to a biochemical or environmental stressor (Buchberger, Yu & Li, 2015; Southey et al., 2014; Yin et al., 2011). It should be noted that proper handling and separation of the samples are key to acquiring quality data, especially in the case of specialized MS techniques such as in vivo sampling methods and MS imaging (Buchberger, Yu & Li, 2015; Gemperline, Chen & Li, 2014; Li, Zubieta & Kennedy, 2009; OuYang, Liang & Li, 2015). Overall, MS provides an attractive ability to examine the full complement of neuropeptides qualitatively and quantitatively.
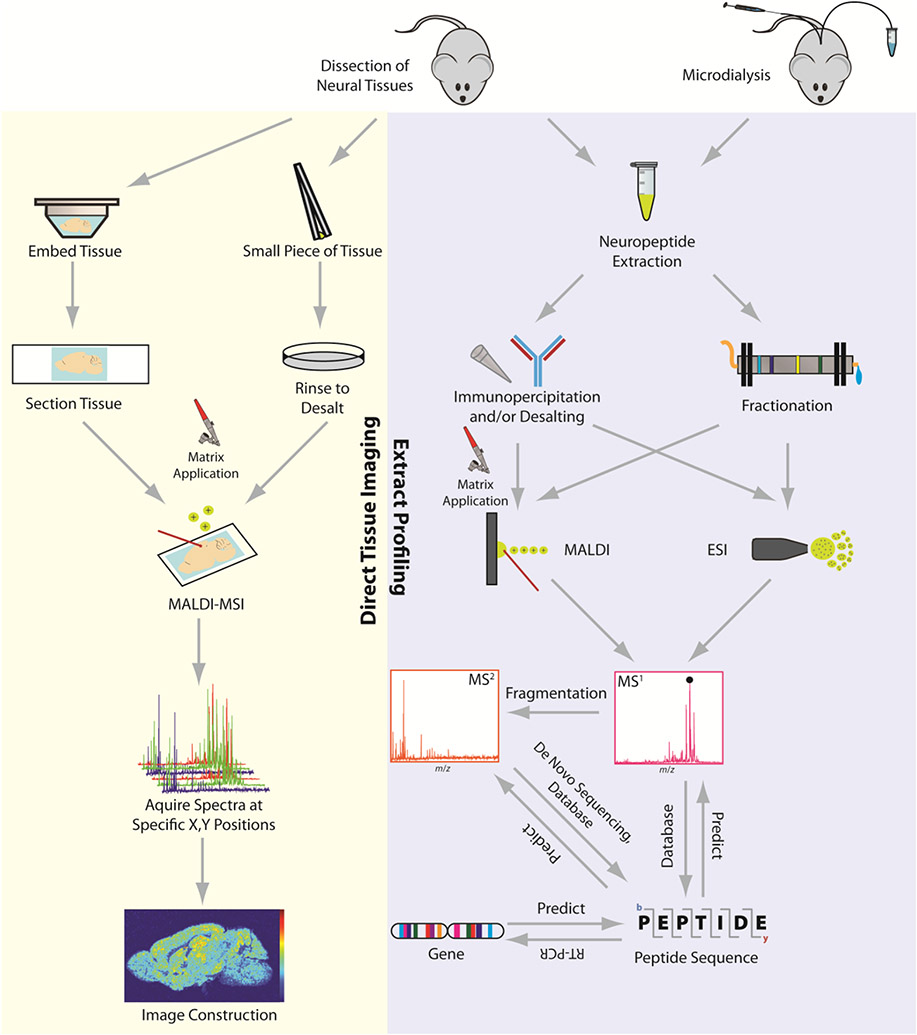
While it seems that MS provides all the necessary qualities to study neuropeptides, many of the techniques used are still far from perfect. Figure 1 provides a pictorial representation of the possible workflows taken when studying neuropeptides with a mouse used as a model organism. This review will focus on the technological advancements and discoveries made, along with the challenging areas that still need development.
Figure 1. General workflow and strategies for investigating neuropeptides by mass spectrometry.
Two major routes: extract profiling and tissue imaging.
Sample Preparation
Sample handling is the first step where researchers need to be cautious to be accurate and consistent. Neuropeptides are often present at low abundance in a background containing all sorts of contaminants (e.g. salts, lipids). They are prone to proteolytic degradation, so sample handling is crucial. Yet it is often the least optimized step compared to down-stream well-established instrumental MS methodologies (Buchberger, Yu & Li, 2015; De Haes et al., 2015; Romanova & Sweedler, 2015; Yu et al., 2014). While salts and lipids compete with neuropeptides for ionization and suppress peptide signals, proteolytic degradation or other protein-modifying enzymes can rapidly change composition of the neuropeptidome, leading to inconsistent and sometimes confounding results.
A. Prevention of Neuropeptide Degradation
Neuropeptides are subject to rapid proteolysis at room temperature. To preserve neuropeptide integrity, flash-freezing of the tissue is convenient to use and widely applied (Han et al., 2015; Sterkel et al., 2011). Other options do exist, including boiling (Altelaar et al., 2009; Sturm, Dowell & Li, 2010; Zhang et al., 2018b) and microwave irradiation (Wardman et al., 2010). Heat denaturation, which was introduced to eliminate post-mortem degradation, can be adapted for a wide range of tissues (Svensson et al., 2009) and has been proven effective. Colgrave et al. have shown that hypothalamic tissue treated with a stabilization device yielded twice the number of mature neuropeptides than those detected in the untreated samples (Colgrave et al., 2011). The Stabilizer T1 (Denator, Gothenburg Sweden), heat stabilization system has been shown to successfully increase neuropeptide identifications compared to other tissue preservation methods. It is worth noting that a high number of identifications may not indicate successful prevention of neuropeptide degradation, but rather abundant peptide signal may be due to high levels of post-mortem degradation (Fridjonsdottir et al., 2018; Yang et al., 2017). Protease inhibitors also serve a similar purpose; for example, Onorato et al. recently showed that recovery of neuropeptide (Pyr)1 apelin-13 from blood samples was only observed when samples were treated with a stabilization cocktail consisting of HALT® protease inhibitor (ThermoFisher Scientific), 0.25 mM phenylmethanesulfonyl fluoride and 25% guanidine HCl (v/v) (Onorato et al., 2019). Protease inhibitors are also added to biological liquids, such as crustacean hemolymph (Chen et al., 2009b).
B. Extraction Strategies
Several workflows exist depending on the type of information sought from the sample (Buchberger, Yu & Li, 2015; Dallas et al., 2015; Yu et al., 2014). Tissue homogenization and peptide extraction are procedures that affects identification rate in neuropeptidomics. Homogenization typically employs manual tissue grinding (i.e., using a pestle on snap-frozen tissue), sonication, or cell disrupter devices. Homogenization and extraction are performed in the presence of solvents or buffers which can dissolve peptides and simultaneously deactivate proteases in the sample. One of the most generally utilized buffers in such application is acidified methanol (Adamson et al., 2016; Budamgunta et al., 2018; Chen et al., 2010c; Hui et al., 2013; Lavore et al., 2018; Sterkel et al., 2011; Van Bael et al., 2018b; Ye et al., 2015) which contains 90% methanol (MeOH), 9% glacial acetic acid, and 1% water. It is reported to be able to extract neuropeptides from single neurons (Zhang et al., 2018a). This buffer system is further optimized by Zhang et al. for a ‘mixing on column’ protocol, an approach that includes four steps with varying aqueous and methanol compositions. This hybrid protocol was able to capture hydrophobic peptides as well as hydrophilic peptides simultaneously and create up to five-fold more neuropeptide identifications (Petruzziello et al., 2012; Yu et al., 2015b; Zhang et al., 2012a). Chen et al. also demonstrated that the use of acidified methanol with a protease inhibitor additive is efficient for trace-level neuropeptide analysis in hemolymph samples (Chen et al., 2009b). However, C-terminal methylation, an enzyme-assisted extraction artifact, might happen to some neuropeptides (Stemmler et al., 2013). Although use of acidified methanol is prevalent, a 0.25% acetic acid solution (DeAtley et al., 2018; Fridjonsdottir et al., 2018) has been shown to produce higher quality neuropeptide signal than acidified methanol (Dowell, Heyden & Li, 2006). Therefore, examples of effective strategies are to perform either multiple peptide extractions on the same tissue homogenate (Petruzziello et al., 2013), collect, and combine the supernatant fraction from each extraction (Yang et al., 2017; Yang et al., 2018). Alternatively, a peptide extraction can be followed by the addition of salt to the peptide extract supernatant to further precipitate remaining proteins (Gomez-Ramos et al., 2018).
A wide variety of organic buffers have been used in the recent years to extract neuropeptides from several biological matrices using acids such as formic acid (FA), trifluoroacetic acid (TFA), and ethylenediaminetetraacetic acid (EDTA), summarized in Table 1. For tissues that are difficult to homogenize, like bone tissue, more corrosive extraction buffers such as 1.2 molarity (M) hydrochloric acid (HCl) and 20% acetonitrile (ACN) are necessary (Gatenholm et al., 2019). Additionally, delipidation strategies using n-hexane (Van Bael et al., 2018a) or methyl-tert-butyl ether (MTBE) in MeOH (Li et al., 2020b) can also be applied during neuropeptide extraction. In lieu of organic solvent extraction buffers, molecular weight cut-off (MWCO) filters have also been used for neuropeptide purification and isolation of a particular size of neuropeptides. For example, neuropeptides from sea cucumber radial nerves can be extracted using either artificial sea water (Chieu et al., 2019a) or simple urea-based cell lysis buffers (Chen et al., 2019) followed by MWCO filters. However, extra care must be taken using these methods to avoid peptide degradation by catabolic enzymes. For biological samples containing abundant high molecular weight proteins, such as hemolymph (Fredrick & Ravichandran, 2012), a combination of extraction using acidified methanol and ultracentrifugation through MWCO filters are necessary for neuropeptide analysis (Liu et al., 2019).
Table 1.
Examples of various organic buffers applied for neuropeptide extraction from different biological material.
| Biological Material | Extraction Buffer | Reference(s) |
|---|---|---|
| various | 90% MeOH, 9% HOAc, 1% water | (Adamson et al., 2016; Budamgunta et al., 2018; Chen et al., 2010c; Hui et al., 2013; Lavore et al., 2018; Sterkel et al., 2011; Van Bael et al., 2018b; Ye et al., 2015) |
| whole sea anemone | 90% MeOH, 9% water, 1% FA | (Hayakawa et al., 2019) |
| rat spinal cord tissues | 80% MeOH, 10% water, 10% FA | (Tillmaand et al., 2020) |
| mice cecum | 37.5% MeOH, 12.5% chloroform, 50% water | (Keller et al., 2020) |
| starfish | 70% MeOH, 5% HOAc | (Kim et al., 2016) |
| bed bug beetle | 50% MeOH, 1% FA | (Predel et al., 2018) (Ragionieri & Predel, 2020) |
| stick insect tissues | 50% MeOH, 1% TFA | (Liessem et al., 2018) |
| various | 0.25% HOAc | (DeAtley et al., 2018; Dowell, Heyden & Li, 2006; Fridjonsdottir et al., 2018) |
| dog saliva human plasma | 80% ACN | (Wang, Marti & Anderson, 2019) (Kirwan et al., 2018) |
| oyster ganglia | 90% ACN, 0.1% TFA | (Schwartz et al., 2019) |
| monkey plasma and cerebrospinal fluid | ACN | (Lee et al., 2018) |
| citrus psyllid colonies | 10% trichloroacetic acid, 2% 2-mercaptoethanol in acetone | (Fleites et al., 2020) |
| sea urchin | 60% acetone, 40% water, 1% HCl | (Monroe et al., 2018) |
| Aplysia abdominal ganglia | 80% acetone, 10% water, 10% FA | (Anapindi et al., 2018) |
| rat spinal cord tissues | dry ammonium sulfate with 0.01 M EDTA | (Do et al., 2018b) |
| bone | 1.2 M HCl in 20% ACN | (Gatenholm et al., 2019) |
C. Enrichment and Sample Clean-Up
Generally, crude neuropeptide extract still contain soluble contaminants, such as salt, which can degrade mass spectral quality and result in decreased peptide signal in MS measurements (Constantopoulos, Jackson & Enke, 1999), and desalting neuropeptide extract is especially important for biological samples that are suspended in proteomics/peptidomics buffers. Examples of popular commercial methods for desalting neuropeptide extract typically involves solid phase extraction utilizing reversed phase resin (i.e., C4, C8, or C18), such as Millipore ZipTip pipette tips (Sigma Aldrich) and Pierce Desalting Columns or Tips (Thermo Fisher), or a hydrophilic polymer sorbent, such as Oasis HLB (Hydrophilic-Lipophilic-Balanced) cartridges (Waters). These types of tools are critical for peptidomics workflows because they not only desalt, but also concentrate neuropeptide samples. Additionally, pooling several tissues, organs, and neurons into one sample is often necessary when concentrated neuropeptidomic content is desired for comprehensive neuropeptide identifications. Other methods of concentrating neuropeptides are by utilizing monoclonal antibodies immobilized on magnetic beads (Vocat et al., 2020), automated solid-phase extraction (Bardsen et al., 2019), and large volume sample stacking using capillary electrophoresis (CE) (DeLaney & Li, 2019a). A recently developed technique to quickly concentrate and desalt neuropeptides involve dispensing a droplet of tissue extract onto a sample target consisting of a hydrophobic circle surrounded by a hydrophilic ring, which allows separation between salts and neuropeptides to occur directly on the MS sampling plate (Wang et al., 2017; Yoon et al., 2018).
D. MS Imaging
Unlike tissue homogenization, direct analysis of intact tissue is a simpler way that enables comparing localization from individual samples or animals which is usually important to determine its biological relevance. For example, intact somata were analyzed after aspiration by a pipette and transfer onto an MS sampling plate for neuropeptide profiling (Diesner, Predel & Neupert, 2018; Neupert et al., 2018). Liquid extraction surface analysis (LESA) is a direct tissue sampling technique that has recently been commercialized by HTX Technologies as the SepQuant droplet probe and has been used successfully for neuropeptide analysis (Kertesz et al., 2015). Pioneered by Caprioli and co-workers, MS imaging has also emerged as an attractive technology for localizing neuropeptides (Caprioli, Farmer & Gile, 1997). Neuropeptide MS imaging experiments require sectioning tissue into 10–20 μm thick slices. Tissues have to be embedded into scaffold materials, such as gelatin (Chen et al., 2010a; OuYang, Chen & Li, 2015; Ye et al., 2015), sucrose (Verhaert et al., 2010), gelatin containing sodium salts of carboxymethyl cellulose (CMC) (Resetar Maslov et al., 2019), at the time of snap-freezing to facilitate sectioning and preserving tissue integrity. Once sectioned, the tissue can be directly mounted onto a glass slide or sample plate for matrix application with an airbrush or automatic matrix sprayer (Andersson et al., 2008; Ye, Greer & Li, 2012). Spectral quality can be improved by washing the tissue sections with organic solvents (e.g. ethanol, methanol, acetone, water, or different mixtures of these solvents) prior to matrix application to remove salts and lipids which negatively influence the matrix crystallization process and signal quality (Buchberger et al., 2020c; Kaletaş et al., 2009; Meriaux et al., 2011; Seeley et al., 2008). The most common matrices for neuropeptide imaging include α-cyano-4-hydroxy-cinnamic acid (CHCA) (Chen et al., 2009b; Pratavieira et al., 2014) and 2,5-dihydroxybenzoic acid (DHB) (Chen et al., 2010a; Ye et al., 2015; Zimmerman et al., 2009). More details on MS imaging are described in a separate section below.
E. Microdialysis
Though tissue homogenization and direct tissue analysis are complementary in gaining insight into sample composition and localization, they all require sacrificing animals. This makes it impossible to track real-time change in vivo and brings in unwanted variations among animals if following time course changes is the real objective. As an emerging as well as underdeveloped technique, microdialysis offers the capability to monitor spatio-temporal dynamics of neuropeptides over a certain time period upon external stimulus via a probe implanted into the tissues of interest that allows continuous sampling from the extracellular space (Kushikata & Hirota, 2011). When sampling from extracellular space, the concentration gradient drives the analytes to diffuse across the dialysis membrane, which has a certain MWCO filter depending on the substances of interest (OuYang, Liang & Li, 2015). Due to the small probe size, animals endure minimal physical damage and associated neurological disturbance. Long-term sampling can be accomplished while animals are still alive and freely moving. It has found its applications in a wide variety of tissues and organs, including skin (Baumann et al., 2019), hypothalamus (Guzman-Ruiz et al., 2015; Kurian et al., 2015), hippocampus (Takeda et al., 2011), spinal cord (Wu et al., 2015b) and kidney (Wesson, Jo & Simoni, 2015) in vertebrates as well as neuronal organs in crustaceans (Behrens, Chen & Li, 2008; Jiang et al., 2016; Liang, Schmerberg & Li, 2015; Schmerberg, Liang & Li, 2015).
Despite its attractiveness, challenges still exist for microdialysis sample preparation. High temporal resolution (shorter intervals for collection of individual samples) is desired for microdialysis measurements, but this must be considered with MS sensitivity factor by selecting an appropriate sampling volume. Balancing low neuropeptide concentration in vivo (1–100 pM), small sample volumes generated by microdialysis (1–10 μL) (Zhou et al., 2015) and low recovery rate (20-30%) (Schmerberg & Li, 2013) makes the choice of instrument even more important. It has been demonstrated that adding organic solvents, especially ACN, to dialysate is able to prevent adsorptive loss of low-abundance neuropeptides by hydrophobic interactions with membrane surfaces (Maes et al., 2014; Zhou et al., 2015). By treating the dialysis membrane and fused silica tubing with polyethylenimine (PEI), recovery was improved by 1.2- to 80-fold (Zhou et al., 2015). This only benefited the detection of peptides that carried a net positive charge, though, probably due to reduced electrostatic interaction between peptides and the microdialysis probe. An array of affinity-enhanced microdialysis approaches have been tested by Schmerberg et al., and they observed antibody-coated magnetic nanoparticles to provide the greatest enhancement in neuropeptide recovery (Schmerberg & Li, 2013). Other efforts to increase peptide recovery include a study by Wanseele et al., who tested several liquid chromatography (LC) columns and mobile phases to find the combination for optimal recovery of neuropeptides (Cortecs®C18+ column with a mobile phase containing methanol as organic modifier and acetic acid as additive) from microdialysate of a solution containing peptide standards (Van Wanseele et al., 2017). Another advancement in microdialysis probe sampling include non-specific perturbing of the tissue of interest to elicit a biochemical response. Al-Hasani et al. developed a microdialysis probe containing optical fibers for the purpose of stimulating neuronal peptide release which is subsequently collected in the probe perfusate (Al-Hasani et al., 2018). The peptide profile resulting from non-specific techniques such as this can be used to generate additional research questions that can be answered by more specific techniques, such as expression knock-out experiments. Although microdialysis is useful for performing in vivo experiments, the recovery rate of neuropeptides is relatively low.
Overall, each sample handling step strives to increase neuropeptide signal by decreasing interfering signal while minimizing sources of neuropeptide loss. However, the variety of chemicals and solvents used by different research groups (even for similar tissue types) illustrates the need for continued evaluation and comparison between these different extraction and sampling systems. Ideally, there would be a workflow that is unanimously agreed upon to produce optimal neuropeptide signal, but it is our opinion that there would likely exist multiple workflows tailored for individual sub-classes of neuropeptides and specific underlying questions to address.
Discovery/Sequence Identification
A. MS in General Peptide Structural Elucidation Strategies
Prior to the introduction of MS, neuropeptides were identified during searches for endogenous molecules that produced a physiological effect, and Edman degradation was used as a standard method to determine the primary sequences (Yu et al., 2015a). This strategy requires a substantial amount of sample, especially from tissue types with scarce neuropeptide content, and a priori knowledge of the analyte of interest since it is a “function first” approach. With its high-throughput capability, MS, especially when coupled with electrospray ionization (ESI) sources, allows thousands of peptides to be measured simultaneously. One of the pioneers in the field, Dominic Desiderio, demonstrated the utility of MS for endogenous (neuro)peptide structural analysis early on (Desiderio et al., 1993; Desiderio & Yamada, 1982; Kusmierz & Desiderio, 1992; Mahajan & Desiderio, 1978; Yamada & Desiderio, 1982). By alternating between MS and MS/MS, records of both intact mass and fragment information (to determine the sequence) can be obtained. Matching these two pieces of information to the respective genome reveals exact neuropeptide sequences, their origins, as well as functions. However, not all organisms have their genome fully characterized, which sometimes makes genomic-based database searching unfeasible. This is overcome by de novo peptide sequencing, a technique that can provide neuropeptide sequences solely based on tandem MS data, without the need for a complete genome. MS has greatly shifted discovery of neuropeptides from the identification of a single peptide to the characterization of multiple peptides representing entire peptidomes.
Various fragmentation techniques have been developed, see Table 2. Collision-induced dissociation (CID), the conventional vibrational activation, has been widely used (Ye et al., 2013; Zhou, Mabrouk & Kennedy, 2013). However, CID has been criticized for preferentially cleaving the weakest bonds, no matter of location in the peptide backbone or side chains, such as with PTMs. Once a bond is cleaved, the internal energy is released and the product will not be further activated, which sometimes leaves spectra with few dominating peaks to interpret (Medzihradszky & Chalkley, 2015; Seidler et al., 2010). Furthermore, the loss of PTMs can be detrimental to some studies. To generate a better-quality spectrum, an alternative fragmentation approach is the beam-type CID or high-energy collision dissociation (HCD). It accelerates all ions across the chamber instead of the ion trap, permitting multiple collisions, and therefore fragments might break up further to create products equally distributed along the backbone (Jedrychowski et al., 2011; Medzihradszky & Chalkley, 2015). Fragmentation by CID in a triple quadrupole and HCD in an Orbitrap mass analyzer for the structural characterization of neuropeptide receptor antagonists were compared (Silva et al., 2018). Similar qualitative and structural information was seen between the two mass analyzers, though higher confidence structural assignments were seen from the HCD-obtained data (Silva et al., 2018). Another comparison was performed by Tu et al. between HCD in an orbitrap, HCD in an ion trap, and CID in an ion trap using an Orbitrap Fusion Lumos where they achieved the highest number of identifications using HCD in the orbitrap, then using HCD in the ion trap, and the lowest amount from CID in the ion trap (Tu et al., 2016). Despite being less sensitive than CID due to the higher ion volume requirement to generate a spectrum, HCD has become more and more popular due to its better data quality and ability to record all products across a wide mass range (Silva et al., 2018).
Table 2.
Four common ion activation and dissociation techniques - CID, HCD, ETD, ECD, and EThcD - for fragmenting peptides.
| Method | Mass Analyzers | Fragment Ions | Advantages | Disadvantages |
|---|---|---|---|---|
| CID | Quadrupole ion trap, triple quadrupole | b-type, y-type | Fast, sensitive, more efficient for low charge ions, induces glycan fragmentation | Low mass resolution, loss of labile modifications |
| HCD | Orbitrap | b-type, y-type | Fast, high mass resolution, induces glycan fragmentation | Less sensitive and slower than CID, loss of labile modifications |
| ETD | Ion trap, ion trap-orbitrap hybrid | c-type, z-type | Retains neutral and labile modifications, faster than ECD, suitable for higher charge state ions (>2+) | Slow, inefficient fragmentation for low charge state precursors, less accessible instrumentation |
| ECD | FTICR or selected time-of-flight | c-type, z-type | Retains neutral and labile modifications | Less accessible instrumentation |
| EThcD | Orbitrap | b-type, y-type, c-type, z-type | Rich sequence-specific fragment ion information, more suitable for characterization of neuropeptides with labile PTMs | Longer duty cycle, requires the production of higher charge state of precursor ions |
Two MS/MS methods complementary to collision-based activation that have been developed are electron-capture dissociation (ECD) (Zubarev et al., 2000) and electron-transfer dissociation (ETD) (Syka et al., 2004), where a radical ion is formed and undergoes fragmentation to yield almost exclusively peptide backbone fragmentation, thus preserving labile PTMs. Following their introduction, both electron-based methods, particularly ETD, have been gaining popularity among researchers studying PTMs in proteomics (Sobott et al., 2009). While still relatively new, ECD and ETD have the potential to be a critical component of neuropeptide sequencing. Unlike the digested protein fragments observed in bottom-up proteomics with predictable C-termini and similar lengths, neuropeptides tend to have varying sizes from a few to several dozens of residues. For example, some FMRFamides in invertebrates have only four amino acids whereas CCK-58, as indicated by its name, has 58. Furthermore, endogenous proteolytic processing leads to the production of peptides containing multiple internal basic residues (histidine, lysine, and arginine) which hold higher charges states in the gas phase, for which CID and HCD show limited performance. Fortunately, that is where ETD outperforms the former two (Hui et al., 2011). Combining CID or HCD with ETD provided complementary spectra for Sasaki et al. in their study on endogenous peptides from a human endocrine cell line, and ETD helped identify a previously unknown large peptide, VGF[554–577]-NH2 (Sasaki, Osaki & Minamino, 2013). Rathore et al. developed a strategy to perform two dissociation techniques, CID and ETD, in one analysis without a decrease in duty cycle. Facilitated by the temporal separation gained through ion mobility MS (IM-MS) (see Isobaric PTMs section), a single packet of precursor ions can give rise to b- and y-type ions containing spectra and c- and z-type ions containing spectra (Rathore, Aboufazeli & Dodds, 2015). A hybrid strategy was further developed by Hui et al. and Jia et al. where a bottom-up approach using CID and HCD fragmentation was coupled with a top-down strategy employing ETD fragmentation to reveal more structural details of large neuropeptides (Hui et al., 2011; Jia et al., 2012). This represents a new route to discovery and characterization of large neuropeptides since neither of these fragmentation techniques could manage to provide a complete picture of a large neuropeptide alone. Rather than using CID, HCD, or electron activated dissociation (ExD), Vrkoslav and colleagues have shown that in-source decay fragmentation can be used to produce fragment ions for peptide structure characterization in single-stage matrix-assisted laser desorption/ionization (MALDI) instruments lacking precursor ion-selection capabilities (Vrkoslav et al., 2018). To improve the coverage and quality of neuropeptide sequencing by in-source decay, Neupert reports a method for N-terminal derivatization using 4-sulfophenyl isothiocyanate (Neupert, 2018). This radical based dissociation technique enables the fragmentation of intact peptide ions, where traditional dissociation techniques are inefficient.
Chemically-derivatized peptides can carry some distinct fragmentation patterns and/or improve fragmentation, and some of them can be utilized for sequencing. Dimethyl labeling is one of the well-established methods that has been employed in neuropeptide identification studies (Fu & Li, 2005; Hsu et al., 2005; Ma et al., 2009), as it features enhanced a1-ion signal for N-terminal determination and simplified MS/MS interpretation. Dimethyl labeling is also effective for analyzing dipeptides and tripeptides (Tang et al., 2014). Short neuropeptides (2-3 residues) are difficult to characterize. They have low molecular weights, complicating the desalting process, and can be hydrophilic, decreasing compatibility with conventional C18 columns. However, these short peptides are still bioactive and potentially important; their MS analysis benefits from derivatization with Marfey’s reagent (Bobba, Resch & Gutheil, 2012). Acetylation is another example of methods that target primary amines (Yew et al., 2009). A nanosecond timescale photochemical click-chemistry based enhancement for neuropeptide detection was developed by Li et al. to remove matrix components to decrease matrix effects and spectral complexity (Li et al., 2019). A few other chemical derivatization schemes have been developed in recent years but have yet applied to neuropeptide studies. Kim et al. reported an oxazolone chemistry for incorporation of Br signature to the C-terminus, which populates MS/MS spectra with a series of y-ions bearing a Br signature for easier interpretation (Kim et al., 2011a). Isothiocyanate analogues with basic moieties have been demonstrated to derivatize peptides and significantly improve the MS sensitivity, while promoting Edman-type cleavage and maintaining other sequence fragments for easy sequencing (Wang, Fang & Wohlhueter, 2009). Cationization by alkali metals have also been shown to improve de novo sequence coverage of small peptides (<15-20 residues) (Logerot & Enjalbal, 2020). The peptide derivatization strategy reported by Frey et al. appends tertiary or quaternary amines to the peptide’s carboxyl groups present at the C-terminus and in aspartic and glutamic acid side chains. As the amine appended, the charge state of that peptide increases, improving its ETD fragmentation efficiency (Frey et al., 2013). Charge state manipulation and distribution of neuropeptides were further studied by Nielsen and Abaye where it was found that the use of electrolyte additives or supercharging reagents was sufficient to alter the observed charge states and total ion signal (Nielsen & Abaye, 2013). Bongaerts et al. recently studied the use of several supercharging agents on neuropeptide ionization and concluded the effects to be highly dependent on the peptide (Bongaerts et al., 2020). While supercharging agents can alter charge state distributions to something more desirable, care must be taken to choose the appropriate one for each analyte.
B. Data Independent Analysis
While improvements in fragmentation techniques have paved the way for the increased identification and characterization of neuropeptides, traditional discovery/shotgun proteomics strategy using data-dependent acquisition (DDA) is still limited by the number of MS/MS spectra abled to be collected. This is problematic for the analysis of more complex samples because only a small fraction of analytes can be selected and fragmented. As the most abundant precursor ions are selected for fragmentation, DDA biases detection to higher abundance or more readily ionizable species. Data-independent acquisition (DIA) can address some shortcomings of DDA, expanding proteome and peptidome coverage through its increased MS acquisition abilities (Chapman, Goodlett & Masselon, 2014). DIA methods involve the isolation and fragmentation of multiple precursor ions within a window simultaneously, with windows spanning the whole m/z range of interest, followed by the use of software to deconvolute the more complicated MS/MS spectra containing fragments from several precursors. This approach generates fragment ions of all precursors in a sample instead of solely the highest abundance ones. The information gathered from every sample component can thus be accessed later as well with the evolution of better software, increasing the capabilities for untargeted analysis. While the additional information enables a wider coverage and increases reproducibility of analysis, the subsequent data deconvolution becomes exponentially more complex. An up-to-date and comprehensive review was written by Zhang et al. addressing several different DIA schemes, as well as software tools for analysis and library building so this will not be addressed again in this review (Zhang et al., 2020a).
While DIA is increasingly being incorporated into proteomics analysis workflows, it is slow to be applied to neuropeptidomics, a field that would benefit greatly from a decrease in high-abundance bias. This is made evident by the work by Kwok et al., where they developed a sensitive method for the detection of 42 bioactive peptides and hormones using DIA (Kwok et al., 2020). A side-by-side comparison performed by Delaney and Li demonstrated the utility and benefits of incorporating DIA over DDA into the neuropeptidomics workflow (DeLaney & Li, 2019b). An impressive improvement was seen in the number of neuropeptide identifications, sequence coverage, and technical and biological reproducibility, further demonstrating the utility of applying a DIA workflow to neuropeptidomics analysis. While it has been demonstrated that a DIA approach can provide benefits over DDA analysis, Saidi et al. also saw an advantage to using parallel reaction monitoring (PRM) to perform targeted peptide quantitation of neuropeptides (Saidi, Kamali & Beaudry, 2019). The authors compared the use of DIA with PRM analysis and observed an increase in variability and decrease in performance associated with DIA, indicating DIA has larger advantages in an untargeted capacity, rather than in targeted analyses. These few explorations into DIA for neuropeptide analyses demonstrate promise for utilizing the advantages of DIA for analysis, though it seems to be slow to be incorporated into the neuropeptidomics workflow, potentially due to a lack of tailored software tools and spectral libraries.
C. Peptide Bioinformatics: Database Search Software/De Novo Sequencing Advances
1. Peptide Sequence Prediction and Databases
Traditional proteomics workflows compare MS-generated fragmentation data to genome-generated databases to determine which proteins are found in a sample. Unfortunately, this workflow does not transfer directly to neuropeptides; a comprehensive specific endogenous (neuro)peptide database does not exist. Several independent initiatives have been initiated in the past. If a species does not have its genome fully sequenced, there is not an easily obtained database to compare against. Furthermore, the fact that neuropeptides go through a series of modifications involving several endopeptidases before final maturation/neuronal release introduces some degree of unpredictability of their final active sequences, meaning that these genomic-generated databases may not be accurate. Therefore, not much can be learned about neuropeptides without robust bioinformatics tools even with a complete genome database. To predict neuropeptide sequences in silico from a genome and construct a reliable database, multiple algorithms have been developed and tested, which has been well-reviewed in several publications (Boonen et al., 2008; Hayakawa et al., 2019; Yu et al., 2014). We have compiled a list of tools and resources, including sequence prediction tools, database compilations, and tools to search MS spectra, specifically developed to benefit the MS identification of neuropeptides in Table 3. Generally, when studying a new organism, the genome of which is available, the online BLAST program allows extraction of all potential neuropeptide prohormones (NPPs) with known NPPs from related species (Christie, 2015; Conzelmann et al., 2013). The deduced NPPs are processed to remove signal peptides using the online program SignalP 5.0 (Almagro Armenteros et al., 2019; Christie, 2015; Petersen et al., 2011), after which they are ready to be submitted to neuropeptide prediction tools such as NeuroPred (Han et al., 2015; Hummon et al., 2003; Tegge et al., 2008), ENPG (Hayakawa et al., 2019), NeuroPred-FRL (Hasan et al., 2021), and specific for insect neuropeptide prediction, NeuroPIpred (Agrawal et al., 2019). Another resource for insect research is DINeR, a database for neuropeptide sequences and functionality (Yeoh et al., 2017). In another homology-based search, Ofer et al. reported a machine learning scheme, Neuropeptide Precursor Identifier (NeuroPID), that can be trained on hundreds of identified NPPs and used to predict metazoan NPPs (Ofer & Linial, 2014). NeuroPP, another tool for neuropeptide precursor prediction has also been developed for improved screening (Kang et al., 2019). Burbach presented an inventory of known neuropeptides, classified in families according to shared structural properties (http://www.neuropeptides.nl) (Burbach, 2010), which is included in another database additionally compiled of genes and precursors called NeuroPep (Wang et al., 2015b). SwePep, while not currently active, was also an endogenous peptide specific database that improved MS analysis (Falth et al., 2006). However useful, these databases are not searchable directly with MS/MS data. NeuroPedia, a specialized neuropeptide database and spectral library that is directly searchable using MS/MS data was constructed, improving identification efficiency, sensitivity, and reliability (Kim et al., 2011b). Instead of using homology-based or de novo sequencing database filtration-based searches, Menschaert et al. developed a genome-wide database searching method combined with de novo sequencing, IggyPep. Compared to using limited-sized database searches, a 30% increase was seen in identification rate when searching the sea urchin neuropeptidome (Menschaert et al., 2010). This approach was later adapted to include enhanced homology-based gene discovery to discover new prohormones and neuropeptides, previously unidentified by the original IggyPep method (Monroe et al., 2018). Also using genomic information, Jarecki et al. discovered novel neuropeptides through searching Ascaris suum libraries of expressed sequence tags and preliminary genome survey sequences (Jarecki et al., 2011). The field of neuropeptidomics faces challenges as many of the model organisms for analysis do not have a fully sequenced genome. To address these informatics challenges, as well as others with endogenous peptide specific concerns in mind, like technical difficulties arising from a lack of enzymatic digestion, a streamlined analytic framework was developed for large-scale peptidomics (Jarecki et al., 2011). By incorporating database mining and predicting fragmentation patterns, many neuropeptides could be identified and 21 putative novel neuropeptides were discovered (Jarecki et al., 2011). Also with the goal of improving endogenous neuropeptide analysis, Secher et al. developed a full workflow, from sample extraction to bioinformatic analysis, for increased identification and insight into function through a prioritization scheme for biologically relevant peptides (Secher et al., 2016).
Table 3.
Various software tools designed specifically for advancing neuropeptide research.
| Type of Tool |
Name | Brief Description |
Link to Resource |
|---|---|---|---|
| Prediction | ENPG | Neuropeptide prediction | https://sourceforge.net/projects/enpg/ |
| NeuroPID | Neuropeptide precursor and neuromodulator prediction | http://neuropid.cs.huji.ac.il/ | |
| NeuroPIpred | Insect neuropeptide prediction | https://webs.iiitd.edu.in/raghava/neuropipred/ | |
| NeuroPP | Neuropeptide precursor prediction | NA | |
| NeuroPred | Neuropeptide prediction | http://neuroproteomics.scs.illinois.edu/neuropred.htm | |
| NeuroPred-FRL | Neuropeptide prediction | http://kurata14.bio.kyutech.ac.jp/NeuroPred-FRL/ | |
| SignalP | Signal peptide prediction | http://www.cbs.dtu.dk/services/SignalP/ | |
| Database | BLAST | Sequence alignment search tool | http://www.ncbi.nlm.nih.gov/BLAST/ |
| DINeR | Insect neuropeptide database | http://www.neurostresspep.eu/diner/ | |
| NeuroPep | Database of neuropeptides, their genes, precursors | http://isyslab.info/NeuroPep/ | |
| SwePep | Endogenous peptide database | NA | |
| MS Data Search | IggyPep | Hybrid de novo and genome wide-database search | NA |
| NeuroPedia | Searchable neuropeptide database and spectral library | http://proteomics.ucsd.edu/Software/NeuroPedia.html | |
| PRESnovo | Motif prescreening prior to de novo sequencing | https://www.lilabs.org/resources |
While not developed specifically for neuropeptide analysis, PEP Search (http://www.mycompoundid.org/mycompoundid_IsoMS/searchSmallPeptide.jsp) (Tang et al., 2014) can be used for the identification of small neuropeptides, such as dipeptides and tripeptides. Besides specialized endogenous (neuro)peptide search engines, common proteomics database search programs can be used to identify neuropeptides, though the translation may not be that straightforward. To provide a reference for people who want to use a common database search program, Akhtar et al. elaborated on the strengths and weaknesses of several of these programs (OMSSA, X!Tandem and Crux) to identify neuropeptides (Akhtar et al., 2012).
2. De novo Sequencing
If genomic information is too scarce to create a thorough NP database, de novo sequencing can be used to derive amino acid sequences of peptides solely based on MS/MS fragmentation spectra. Since the late 1990s, a handful of de novo sequencing tools have been developed (e.g. PEAKS, PepNovo). A more comprehensive review of de novo sequencing tools can be found in other reviews (Allmer, 2011; Ma & Johnson, 2012). As high resolving power and accuracy are extremely important when deriving a peptide sequence, modern mass spectrometers will continue to make de novo sequencing easier with instrumental advances, which in turn requires new de novo sequencing software tools to be developed accordingly to work with certain type of instruments. For example, pNovo was designed for use with HCD fragmentation (Chi et al., 2010). UniNovo was introduced two years later, claiming to be able to work well for spectra from various types of fragmentation methods (CID, ETD, HCD and CID/ETD) (Jeong, Kim & Pevzner, 2013). Later, Ma et al. presented a novel de novo sequencing program, Novor, offering improvements in both the speed and accuracy for peptide de novo sequencing analyses (Ma, 2015), compare to PEAKS (Mazurais et al., 2015). Most recently, DeepNovo was introduced by Tran et al., an innovative deep learning-based approach for de novo sequencing, outperforming PEAKS, PepNovo, and Novor (Tran et al., 2017). This method was later adapted to create DeepNovo-DIA for analyzing DIA data (Tran et al., 2019). While not created for endogenous peptide analysis, the field of neuropeptidomics benefits from incorporation of these tools into the neuropeptide analysis workflow.
Neuropeptide identification has been facilitated by these various advances and can be further improved through preliminary processing prior to database searching. PRESnovo was developed to take advantage of the common conserved sequence motifs found in many neuropeptides as a prescreening method to improve the subsequent de novo sequencing (DeLaney et al., 2020). By searching through a predefined motif database, probable motifs can be assigned to each precursor from a MS/MS spectrum, which increases correct identifications seen through PEAKS, compared to without PRESnovo prescreening (DeLaney et al., 2020). Preprocessing was also shown to be beneficial for the detection of neuropeptides, using a MATLAB-based workflow and statistical analysis (Salisbury et al., 2013).
After receiving the results from a database search, the confidences of identifications must be evaluated, commonly using statistical false discovery rates (FDRs) and dummy databases (Jeong, Kim & Bandeira, 2012). This is important for measuring the integrity and confidence in identification assignments. Using a mixed species database, the assignment fidelity and false positive percentages were compared after the acquisition of single species neuropeptidomic data using Orbitrap, ion trap, and quadrupole time-of-flight (TOF) instruments (Anapindi et al., 2018). While all platforms saw a decrease in identifications during the use of the mixed database, the Orbitrap data was least negatively affected (Anapindi et al., 2018). Overall, the quantity, quality, and reliability of neuropeptide identifications depends on the careful consideration of neuropeptide sequence prediction, database selection method, as well as the search method and fidelity evaluation parameters. While there are various tools available for identification (and possibly support quantitative analysis), these software are not created for the characterization of endogenous peptides specifically; instead, modern day software requires researchers to state that no enzyme digestion is performed. The field of neuropeptidomics could benefit from development of effective bioinformatic tools able to perform identification without specification of an enzyme or able to interpret results at the endogenous peptide level, rather than having to compromise and use the “digested peptides” function at the software-designated protein level.
Structural Analysis
A. Post-Translational Modifications
As described above, neuropeptide synthesis begins with a large precursor protein that undergoes cleavage by proprotein convertases. These processed peptides are subject to various PTMs, all of which can affect neuropeptide binding affinity, lifetime, and function (Hokfelt et al., 2000). PTMs along with proteolytic processing leads to the generation of distinct structures of bioactive peptides. Such PTMs, such as phosphorylation, sulfation, and glycosylation, may be introduced prior to or after proteolytic processing. While studies to determine the presence of PTMs are important, it is also of interest to understand the mechanisms for modification of neuropeptides (Hook et al., 2018). Location of a PTM, whether on the precursor peptide or on the bioactive peptide, may also be of importance. Multiple prolactin variants were recently identified and their regulation patterns were found to differ (Qian et al., 2018). Glycosylation of the mature natriuretic peptide hormone family alters processing, whereas the O-glycosylation of the propeptide decreases cleavage frequency and leads to fewer bioactive peptides in circulation (Hansen et al., 2019). In addition to the effects from propeptide modifications, altered receptor activation and increased stability of the bioactive peptides were also observed when glycosylation was located on the receptor binding region of the mature peptide (Madsen et al., 2020). Whereas formerly, bioactive neuropeptide PTMs were thought to be conserved to terminal amino acids (for protective effects against degradation) as well as the precursor proteins (for cleavage purposes), though PTMs at other positions along the neuropeptide backbone are likewise observed (Baggerman et al., 2004; Busby et al., 1987; Hummon et al., 2003).
The most common PTMs on neuropeptide termini include pyroglutamate modification of the N-terminus (Gade & Marco, 2015; Lee et al., 2010; Monroe et al., 2018; Salisbury et al., 2013), which is thought to protect the peptide from enzymatic degradation (Hayakawa et al., 2019), and C-terminal amidation, which is required for the biological activity of many neuropeptides (Anapindi et al., 2018; Salisbury et al., 2013; Secher et al., 2016). To evaluate the importance of neuropeptide amidation, Van Bael et al. designed a gene knockout experiment targeting three putative neuropeptide amidation enzymes in Caenorhabditis elegans, an organism able to survive without neuropeptide biosynthesis enzymes. Their findings indicated the dependence on C-terminal amidation for reproduction, drastically interfering with the quantity and success of egg-laying, further highlighting the importance of such PTMs (Van Bael et al., 2018b).
Another common peptide hormone PTM is acetylation. Biological roles of acetylation include to increase peptide stability, by protecting the peptide from enzymatic degradation, and to regulate receptor affinity (Van Dijck et al., 2011; Zhang et al., 2012a). During the characterization of pro-opiomelanocrtin related hormones, Yasuda et al. identified novel tri-acetylation of α-melanocyte-stimulating hormone (MSH) (Yasuda et al., 2011). Acetylation has also been found to exist as a tissue specific modification of mouse hemokinin-1, detected only in the brain and not in peripheral tissue, indicating a brain specific functional role for this PTM (Deliconstantinos et al., 2017).
Cysteine disulfide crosslinking of peptides is an important PTM observed in neuropeptides (Jia et al., 2012). It provides structural rigidity and contributes to a peptide’s three-dimensional structure, essential for receptor recognition and peptide function. Challenges in MS analysis of disulfide crosslinked molecules include its low abundance and low fragmentation efficiency, owing to the stability of the disulfide bond. Yu et al. developed a targeted ETD-based method and data mining scheme to improve the recognition and localization of endogenous disulfide bonds in rat neuropeptides, enabling future studies to target this PTM in a more high throughput manner (Yu et al., 2015b). To improve disulfide bond characterization, Bhattacharyya et al. developed DisConnect, an open source software, to determine disulfide connectivity of peptide hormones, peptide toxins, and proteins, and to characterize disulfide foldamers (Bhattacharyya et al., 2013). In-source reduction methods have also been shown to successfully map disulfide bond linkages in peptides (Cramer et al., 2017; Stocks & Melanson, 2018; Stocks & Melanson, 2019; Ye et al., 2015). A vendor neutral software tool, DiSulFinder, was designed to identify peptide backbone fragments with both intact or cleaved sulfur-sulfur or sulfur-carbon bonds (Liang et al., 2018). Liang et al. were able to quickly provide identifications for disulfide linkage determination in the inter-chain disulfide-linked crustacean cardioactive peptide and insulin fragment peptide (Liang et al., 2018).
Glycation, a PTM associated with age, altering protein structure and function, has also been shown to modify neuropeptides. The different types and binding sites of glycation for the neuropeptide substance P (SP) were investigated by Lopez-Clavijo et al. Using a multimodal MS approach, the authors were able to confidently assign binding sites and identify intermediate products to understand glycation and its different types, paving way for studies of glycation on other neuropeptides (Lopez-Clavijo et al., 2012).
Acidic modifications such as phosphorylation of serine, threonine, or tyrosine and sulfation of tyrosine can benefit from the use of negative ion mode MS analysis and are commonly analyzed through such methods (DeLaney, Phetsanthad & Li, 2020). With only a mass difference of 0.0095 Da between the phosphorylation and sulfation modifications, and both capable of modifying tyrosine residues, HRAM instruments must be used to resolve these small differences (for more Isobaric PTMs see the Isobaric PTMs section). Using a high-resolution Fourier-transform ion cyclotron resonance MS (FTICR-MS), tyrosine sulfation was identified and localized during the top-down analysis of a sex ganglia-specific peptide in Hirudo medicinalis. Sulfation was confirmed through high mass accuracy measurements as well as characteristic isotopic abundance shifts consistent with sulfur isotopes (Hsu et al., 2017). In another study, during a multi-MS platform neuropeptidomic characterization of the rat habenular nuclei, novel sulfation sites were discovered on secretogranin I prohormone and confirmed through an additional targeted MS analysis (Yang et al., 2018). In summary, although it is difficult to differentiate between the two PTMs, it can be achieved using the proper MS tools. In addition, enzymatic tools may help conclusively establish sulfation (de Vries et al., 2005). Neuropeptidomics can benefit from the increased characterization of these two PTMs, as there are many sulfated neuropeptides with unknown function (Seibert & Sakmar, 2008).
Furthermore, phosphorylation is known to induce dynamic modifications of neuropeptides and is of great interest for characterization as potential biomarkers because of its common occurrence (Yasuda et al., 2011). Over 50 novel neuropeptide phosphorylation sites were discovered by Secher et al. by a newly developed bioinformatics tool. Functional studies show that phosphorylation of α-MSH reduces its binding to melanocortin receptors. Serine phosphorylation of neuropeptides were of much higher abundance compared to intracellular proteins in the rat brain (Secher et al., 2016). While insect phosphorylated neuropeptides are rare, Sturm and Predel were able to identify phosphorylation of CAPA pyrokinin in Lamproblatte albipalpus that is interestingly taxon specific. Phosphorylation has not been observed in the closely related species Periplaneta americana even though both cockroach share identical neuropeptides sequences. This suggests some specific development within the peptidergic system of L. alibipalpus requiring phosphorylation for function (Sturm & Predel, 2014). As phosphorylation is known to differentially modify neuropeptides in diverse ways due to the dynamic nature of neuropeptides, Lietz et al. did a study to determine a global status of the phosphorylated neuropeptidome of bovine dense core secretory vesicles through characterizing phosphorylation stoichiometry and site motifs of phosphopeptides. Among a wide range of phosphosites detected, SxE was found to be the most prevalent motif (Lietz et al., 2018). They also found differential regulation of neuropeptides, as expected, on many neuropeptides with both known and unknown function, confirming that there is ample room for future studies into the roles of neuropeptide phosphorylation.
B. Glycosylation
Glycosylation is among the most ubiquitous and complex PTMs in biology, with a diverse range of structural possibilities leading to a variety of functional effects. These include an improved metabolic stability to increase peptide hormone circulatory half-life (Flintegaard et al., 2010). There are several types of glycosylation, primarily N-linked and O-linked. Glycosylation micro- and macro-heterogeneity has been observed on hormones and peptide hormones as well, demonstrating the high degree of diversity of glycans able to modify neuropeptides (Bousfield et al., 2015). Glycosylation is also shown to affect neuropeptide receptors (Quistgaard et al., 2014). Cao et al. analyzed the biosynthesis pathway of calcitonin, a peptide hormone implicated in cancer, and discovered O-glycosylated calcitonin. They observed that both hormone forms responded similarly when the cells were challenged with biosynthetic enzyme inhibitors (Cao et al., 2017). This observation demonstrates the diverse range of glycosylation effects, as it has also been shown to alter response to enzymatic activity (Goettig, 2016).
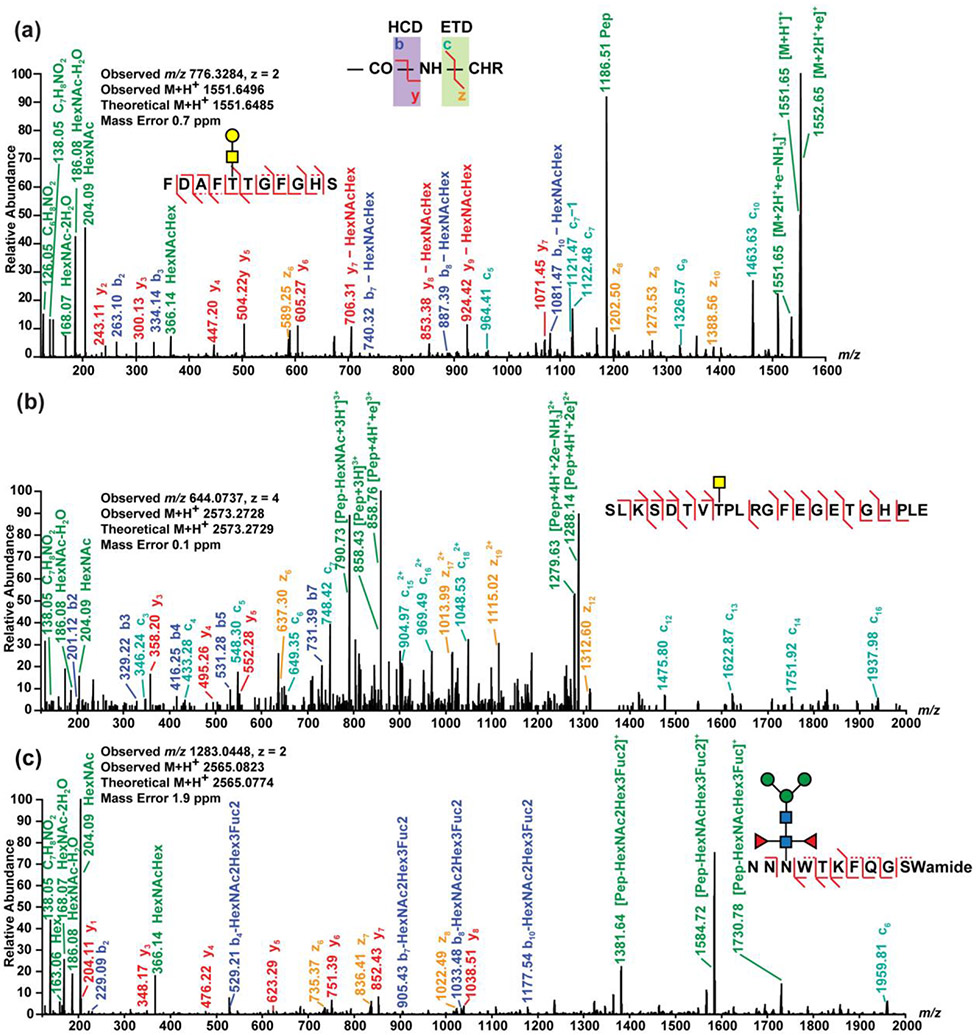
The characterization of glycosylation is important because unlike other simpler PTMs with a static mass shift, glycan composition, as well as the connectivity and configuration of their glycosidic bond, can vary. To increase detection sensitivity and specificity as well as provide improved sequence coverage, Yu et al. used a targeted analytical method employing oxonium ion-triggered electron-transfer/higher-energy collision dissociation (EThcD) (Yu et al., 2017). Demonstrating its utility for neuropeptides modified by glycosylation, several glycosylated signaling peptides were analyzed and several glycoforms were identified. Additionally, novel glycosylated insulin-B chain, insulin-C peptide, and BigLEN, a potential body weight regulating neuropeptide, from mouse and human tissue were reported (Yu et al., 2017). They could distinguish two isobaric monosaccharides, GalNAc and GlcNAc (Yu et al., 2017) through their distinct diagnostic oxonium ion fragmentation profiles (Halim et al., 2014). This targeted method enables higher quality fragmentation spectra to be obtained, along with reducing instrument time required for glycopeptide analysis. Cao et al. also employed oxonium ion-triggered EThcD to characterize both N-linked and O-linked glycosylated neuropeptides in crustaceans (Figure 2) (Cao et al., 2020). In a pursuit to improve the characterization of glycopeptides, Riley et al. systematically compared several fragmentation methods and dissociation energies. The authors found the optimal dissociation methods to differ between N- and O- linked glycans (Riley et al., 2020). While these results were obtained through enzymatically digested peptides, the differences in optimal fragmentation methods likely hold true for endogenous peptides. Thus, the characterization of each type of glycosylated neuropeptides, whether N- or O-linked, should include considerations for each fragmentation method before use.
Figure 2. Using EThcD, both N- and O-linked glycosylated neuropeptides are identified.
EThcD spectra of an (a) O-linked orcomyotropin neuropeptide discovered in rock crab C. irroratus nervous system, an (b) O-linked truncated crustacean hyperglycemic hormone precursor-related neuropeptide discovered in blue crab C. sapidus nervous system, and an (c) N-linked B-type allatostatin (AST-B) neuropeptide discovered in C. sapidus nervous system. Reprinted with permission from (Cao et al., 2020).
Advances in glycoinformatics to aid in glycopeptide characterization include the compilation of several glycomics databases, such as GlyTouCan (Tiemeyer et al., 2017) and glypy (Klein & Zaia, 2019), for glycan identification (Campbell et al., 2014; Ranzinger et al., 2015). There has also been the development of many software programs such as MSFragger (Kong et al., 2017), GlycReSoft (Klein, Carvalho & Zaia, 2018), and O-pair search with MetaMorpheus for O-glycopeptides (Lu et al., 2020). Byonic, a glycoproteomics search program recently added the capability for a glycan “wildcard search” to improve detection of glycans without a priori knowledge of their mass (Roushan et al., 2020). This is beneficial in the neuropeptidomic studies of organisms with incompletely sequenced genomes and lack of knowledge of potential glycans. More detailed information on the glycomics databases and bioinformatics tools available can be found in various platforms (Aoki-Kinoshita, 2017; Dallas et al., 2012; Tsai & Chen, 2017; Woodin, Maxon & Desaire, 2013). A table of useful information for recent MS-based strategies and software tools for glycopeptides is included within a recent review article (Cao et al., 2021).
In a large-scale effort to map O-linked glycosylation on peptide hormones, Madsen et al. found almost a third of the 279 identified peptide hormones to be O-glycosylated, serving as a basis for global O-glyconeuropeptide discovery (Madsen et al., 2020). While peptide hormone glycosylation seems common, it is still of low abundance and is still difficult to detect, let alone characterize and quantify. Thus, several strategies involving enrichment and derivatization schemes were developed. Interested readers are encouraged to examine the reported by Liu et al. for more information on different strategies for the isolation and characterization of glycosylated neuropeptides (Liu, Cao & Li, 2019). Additionally, a comprehensive review about glycopeptide quantitation was published very recently (Delafield & Li, 2020).
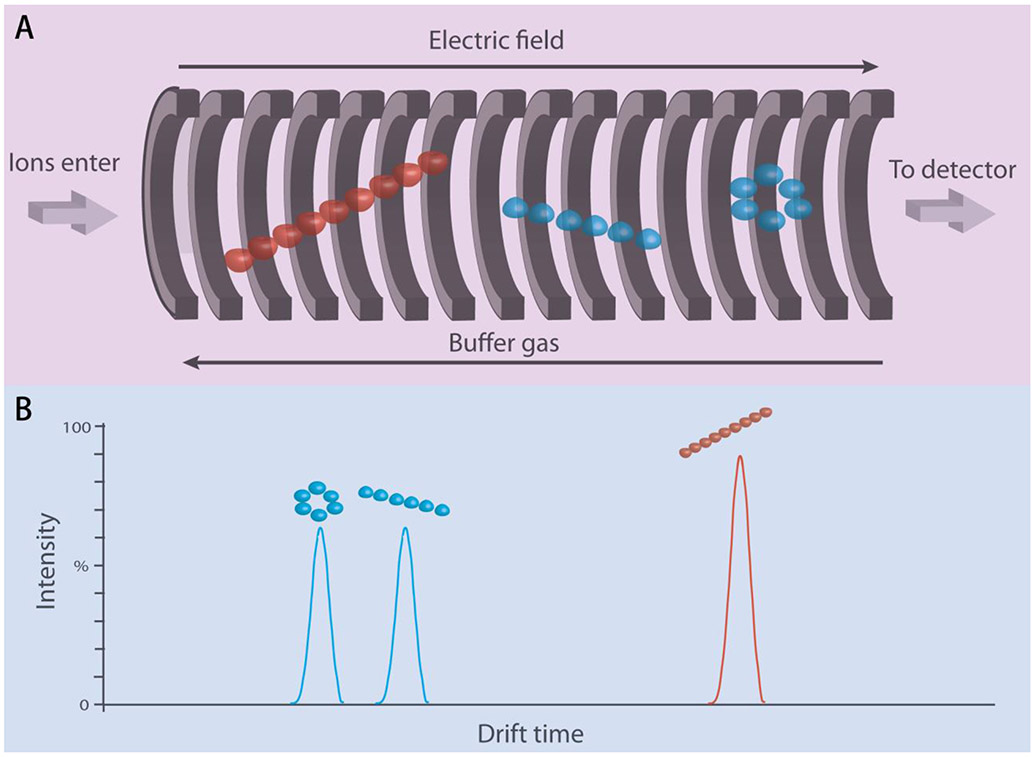
C. Ion Mobility MS
Isobaric species are challenging to study with MS due to their identical nominal masses, especially in a discovery-based mode. Nonetheless, differentiation between different isobaric peptides is important as different isobaric neuropeptides may have different properties and bioactivity. Ion mobility mass spectrometry (IM-MS) is an analytical technique that separates gas-phase ions based on their differences in collisional cross section (mobility) through the buffer gas, which originate from differences in size and shape. The separation mechanism of ion mobility is demonstrated in Figure 3. IM-MS for structure elucidation of isobaric peptides when mass spectrometry measurements is reviewed by Li et al. (Li, Delafield & Li, 2020). Lamont et al. utilized IM-MS and detected two coeluting isobaric peptides, which they identified as the opioid neuropeptides, leucine enkephalin, and N-acetylated alpha-melanocyte stimulating hormone (Lamont et al., 2017). Aspartic acid isomerization to isoaspartic acid is suggested to play a role in apoptosis and protein stability, but the crucial differentiation via MS remains to be challenging. Sargaeve et al. demonstrated the ability to distinguish between these isomers using diagnostic fragment ions produced by ExD fragmentation methods (Sargaeva, Lin & O'Connor, 2011).
Figure 3. Schematic diagrams of analyte ions separation in a drift tube (small ion in blue, large ion in red).
(A) Analytes are ionized and enter the drift tube. Small ions travel faster in the drift tube due to less collision with the buffer gas. In this example, the small ion has two conformations: compact ring conformation and unfolded linear conformation. Same ion with compact conformation will travel faster than unfolded linear species. (B) Drift time profile of analyte ions.
Naturally occurring amino acids in peptides and proteins are typically of the L-isoform, with the D-isoform being rare. Even so, D-amino acid containing peptides (DAACPs) can be found in nature and are the focus of many studies as this “unnatural” stereoisomer can have implications for 3D conformation, bioactivity, and degradation. While many studies have been performed on DAACPs, little of this has been applied to the neuropeptidome. DAACPs can differentially regulate neuropeptide activity by altering affinity to its receptors. Using a combination of IM-MS, computer modeling, cell-based assays and results from prior functional studies (Bai et al., 2013), the Sweedler group discovered and evaluated several analogues of the D-amino acid containing neuropeptides GFFD and GTFD in the sea slug, Aplysia californica (Do et al., 2018a). Careful modeling led to correctly predicting activities with a feeding circuit related receptor, showing the change from L-Ala to D-Ala to alter peptide activity (Do et al., 2018a). The Sweedler group has led many recent efforts in understanding bioactive DAACPs through studying Aplysia californica. One of their workflows analyzes the relative abundances of key chirality-reporting fragment ions to distinguish between neuropeptide L- and D- epimers (Bai, Romanova & Sweedler, 2011). Analyzing single neurons with MALDI tandem MS, identification of D-isoforms of endogenous peptides was demonstrated directly from cells and tissue (Bai, Romanova & Sweedler, 2011). In addition, they evaluated several protocols for untargeted DAACP discovery, again using sea slug neurons (Livnat et al., 2016). Their validated approach involves screening for resistance to aminopeptidase M digestion, inducing a retention time shift between epimers, and comparing the endogenous peptide with synthetic standards leading to the discovery of two peptides with D-isomers. Only one of these peptides appeared to be bioactive (Livnat et al., 2016). For a neuropeptide natively present as both L- and D-residue containing forms, both were found to activate their newly identified receptor, with the D-epimer being the more stable (Checco et al., 2018). The same group also discovered ten new DAACPs in the central nervous system (CNS), two of which were found to be the first animal DAACPs with more than one D-amino acid residue (Mast, Checco & Sweedler, 2020). This demonstrates the dynamic nature of D-isomerization to alter neuropeptides, highlighting the importance of D-epimer localization.
Benefits of DAACPs include enhanced metabolic stability; they are protected from many endogenous enzymes that only recognize the L-amino acid variant. Demonstrating the utility of modified neuromodulators in their exploration for improved pharmacological peptides, Magafa et al. created a variety of neurotensin analogues. Using various combinations of D-amino acids and an unnatural amino acid, they discovered several modified neurotransmitters with improved enzymatic stability, establishing a basis for the rational design of novel pharmaceutical neuromodulators (Magafa et al., 2019). While there have been several method developments for endogenous DAACP detection and identification, specific D-residue peptide localization tends to be complex or expensive (Soyez et al., 2011). Jia et al. demonstrated the utility of a MS fragmentation-based IM-MS method to localize D-amino acid residues in bioactive peptides in a single MS analysis (Jia et al., 2014; Jia et al., 2016). As peptide epimers are chromatographically separated, each can be fragmented by CID prior to ion mobility separation to indicate the presence and location of a D-amino acid. The increasingly known variability and complexities of the effects of this PTM are why DAACPs will retain interest in the future.
D. Conformational Analysis by Ion Mobility MS
While neuropeptides are often considered as 2D entities, it is important to note that these analytes have 3D structures that can widely vary. When combined with molecular dynamics (MD) simulations, IM-MS is able to provide gas-phase peptide ion structural insights at the atomic level. With IM-MS, analyte structure is determined from experimental values measuring temperature-dependent rotationally averaged collision cross sections (CCS). It is hypothesized that these reflect the gas-phase ion conformations originating from solution-phase after desolvation (Jurneczko & Barran, 2011). Compared to other biophysical techniques, such as X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy, IM-MS is sufficiently specific and sensitive to ascertain structural information using impure, trace amount of sample (Scarff et al., 2008). Moreover, whereas X-ray crystallography and NMR provide an averaged structure, IM-MS obtains snapshots of short-lived intermediates and conformational transitional states and thus can be used to interrogate dynamic heterogeneity (Gidden & Bowers, 2002; Gidden, Bushnell & Bowers, 2001). In fact, quite a few studies (Bereszczak et al., 2012; Jenner et al., 2011; Shi et al., 2012; Shi et al., 2014; Wyttenbach et al., 2009) report analyte ion gas phase structure and conformational dynamics, which provides important insights into what occurs in solution.
The conformation of neuropeptides is a very relevant aspect with respect to their biological function. Bradykinin (BK), a nine residue neuropeptide, has been a model peptide both for conformational dynamics studies (Papadopoulos et al., 2012; Pierson et al., 2013; Pierson et al., 2011; Pierson & Clemmer, 2015; Pierson, Valentine & Clemmer, 2010; Voronina & Rizzo, 2015) and the development of systemic IM-MS strategies for structural studies in general. IM-MS assisted by MD revealed 10 independent populations of structures in solution and 3 gas-phase quasi-equilibrium conformations due to combinations of three cis and trans prolines (Pierson et al., 2013; Pierson et al., 2011; Pierson & Clemmer, 2015; Pierson, Valentine & Clemmer, 2010). The Clemmer group investigated penultimate prolines in Substance P (SP). In a detailed and step-by step manner to elucidate the spontaneous peptide cleavage pathway, they showed trans to cis configurational changes to be key in initiating non-enzymatic degradation (Conant et al., 2019). Inspired by the fact that penultimate proline residues are frequently found in neuropeptides, Clemmer and co-workers (Glover et al., 2015) utilized IM-MS to probe the effect of penultimate proline on neuropeptide conformations. Besides protecting peptides from enzymatic degradation, penultimate Pro also plays a key role in increasing the conformational heterogeneity of neuropeptides, which may be important for receptor affinities and thus function.
IM-MS is able to distinguish between cis and trans isomers of Pro-containing peptides (Shi et al., 2016; Warnke, von Helden & Pagel, 2015). While different observed conformations are attributed to the isomerization of proline using specific criteria, they do not always indicate cis/trans conformers; IM-MS has limitations for structure elucidation. To this end, the non-proline containing neuropeptide Y wild type and naturally occurring proline containing mutant were investigated by Lietz et al. Though typical cis/trans isomerization hallmarks were present, the presence of these isomers were excluded (Lietz et al., 2016). IM-MS and MD analyses have their limits and require other methods for validation. While Konig et al. were originally unable to prove DAACP in cicada hypertrehalosemic neuropeptides using IM-MS (Konig, Marco & Gade, 2017), they later showed that other techniques, such as NMR, may be required to confirm the proposed 3D structure (Konig et al., 2019).
Some neuropeptides are active through self-oligomerization (Cowley et al., 1992; Smith & Griffin, 1978) and IM-MS has proven instrumental to study this process. For instance, important insights have been obtained on the amyloid fibril formation that is a central implication in neurodegeneration, including Alzheimer’s or Parkinson’s diseases (Bernstein et al., 2009; Bleiholder et al., 2011). Subsequently, IM-MS studies of various Leu-enkephalin mutants highlighted the importance of characterizing dimer and higher oligomers in determining possible protofibril structures that a peptide system can access (i.e., single β-sheet or doublesheet steric zipper) (Bleiholder, Dupuis & Bowers, 2013; Do et al., 2014).
A number of studies (Heck, 2008; Kaddis & Loo, 2007; Kondrat et al., 2013; Konijnenberg, Butterer & Sobott, 2013; McAllister et al., 2015) demonstrated that certain peptide and protein ions in the gas phase retain a memory of their solution structures upon ionization (e.g., ESI). How exactly the structure in the gas-phase mimics the solution phase remains to be clarified. The Russell group (Fort et al., 2014; Servage et al., 2015; Silveira et al., 2013a; Silveira et al., 2013b) used cryogenic IM-MS (cryo-IM-MS) to reveal that intramolecular interactions can stabilize the kinetically trapped SP dehydrated conformer in a time scale of several milliseconds. The use of cryo-IM-MS for the study of analyte structure and is reviewed by Servage et al. (Servage et al., 2016).
Besides peptide inherent secondary structure, external environmental factors also affect peptide conformational preference in the gas phase. IM-MS studies have investigated temperature (Berezovskaya, Porrini & Barran, 2013; Zilch et al., 2007), activation voltage (Pierson, Valentine & Clemmer, 2010), solvent composition (Pierson et al., 2011), and metal binding (Chen, Gao & Russell, 2011).
MS Imaging
Until recently, the most popular way to obtain spatial information of molecules was immunohistochemistry and RIA, However, due to the selectivity of the antibodies used, immunoassays are unable to acquire information from more than one analyte. This is a disadvantage when working with limited amount of sample. Within the last years, MS imaging (MSI) has emerged as an alternative method to circumvent this disadvantage. Through MSI, molecular ion images are generated of a surface (e.g., tissue or tissue slice). By rastering a laser along a predefined (x, y) grid, a mass spectrum is acquired at each grid square (i.e., pixel). Ion specific images are then generated by bioinformatic tools. As such, MS imaging has the capability to generate hundreds of images from a single experiment. The investigation of neuropeptides and their spatial distribution patterns has been accomplished by MS imaging throughout several organisms for several applications (Altelaar et al., 2005; Berisha et al., 2014; Buchberger et al., 2020c; Chen et al., 2010b; Chen & Li, 2010; Chen et al., 2010d; De Haes et al., 2015; Hanrieder, Ljungdahl & Andersson, 2012; Herbert et al., 2010; Jia et al., 2012; Mark, Maasz & Pirger, 2012; Monroe et al., 2008; OuYang, Liang & Li, 2015; Pratavieira et al., 2014; Romanova et al., 2009; Shariatgorji, Svenningsson & Andren, 2014; Ye et al., 2013; Ye et al., 2015). While the general workflow has become very well defined (Figure 1), several modifications have been explored and implemented to improve the quality and depth of MSI data. Several reviews discuss these in the context of neurobiology (Buchberger et al., 2018; Gemperline, Chen & Li, 2014; Hanrieder, Malmberg & Ewing, 2015; OuYang, Liang & Li, 2015).
A. Ionization, Identification, and Instrumentation
While several ionization techniques exist, only a small subset has been used for MS imaging in biological relevant experiments. MALDI MS imaging (MSI) was first developed by the Caprioli group, who successfully imaged proteins and peptides in thin tissue slices of the rat pituitary and pancreas (Caprioli, Farmer & Gile, 1997). MALDI still remains the most utilized ionization methods for MSI of biomolecules, including metabolites, lipids, and proteins (Eriksson et al., 2013). Alternative ionization methods employed include desorption electrospray ionization (DESI) (Wiseman et al., 2008), nanostructure initiator MS (NIMS) (Sturm et al., 2013a; Yanes et al., 2009), and secondary ion MS (SIMS)(Altelaar et al., 2005; Jiang et al., 2014; Lanni et al., 2014; Monroe et al., 2008; Ogrinc Potocnik et al., 2017), but MALDI MSI has been a predominant technique utilized in neuropeptide-related studies (Buchberger et al., 2020c; Chen et al., 2010b; Chen et al., 2010d; Herbert et al., 2010; Jia et al., 2012; Lanni et al., 2014; Pratavieira et al., 2014; Verhaert et al., 2007; Verhaert et al., 2010; Ye et al., 2013; Ye et al., 2015). Crustacean neuronal tissues have been studied under various MSI conditions to understand the functional roles of neuropeptides through mapping their localization (Buchberger et al., 2020c; Chen et al., 2010b; Chen & Li, 2010; Chen et al., 2010d; Jia et al., 2012; OuYang, Liang & Li, 2015; Ye et al., 2013; Ye et al., 2015). The alternative ionization methods suggest various advantages over MALDI, such as being matrix-free, preventing analyte diffusion (see sample preparation below). They also have distinct disadvantages. For example, while NIMS is excellent for metabolites, lipids, etc., it has been shown to not ionize neuropeptides efficiently when compared to MALDI (Sturm et al., 2013a). For additional information on MS imaging, we refer the reader to an in-depth review on developments in high resolution MALDI MS relevant for neurobiology (DeLaney, Phetsanthad & Li, 2020). For a summary of the advantages and disadvantages of different MS imaging ionization sources (i.e., MALDI, SIMS, NIMS, DESI, and LAESI, and LESA), we refer the reader to Table 1 in a recent review (Rocha, Ruiz-Romero & Blanco, 2017).
With only minor amino acid differences between neuropeptides in the same family, methods for confident identification need to be in place. Classically, MS/MS of singly charged ions, which are primarily produced during MALDI ionization, is inefficient, leading to poor fragmentation and thus inconclusive identifications. Also, due to the varying distribution of analytes across a tissue, the motion of constantly rastering across the tissue makes it difficult to be able to fragment a mass that was originally detected in a previous raster step. Thus, accurate mass matching followed by subsequent tissue extract ESI MS/MS analysis (Ly et al., 2019) have been common ways to identify analytes. With the development of modern instrumentation, such as the HRAM Orbitrap, identification of analytes with similar masses has become more reliable (Verhaert et al., 2010). Yet, tandem MS is still difficult on singly-charged ions. Significant effort has been put in developing hybrid methods of MS and MS/MS occurring in a single square form in order to facilitate the isolation of ions identified in first pass MSI spectra (OuYang, Chen & Li, 2015). These hybrid methods also increase the image quality, as shown in Figure 5 (OuYang, Chen & Li, 2015).
Figure 5. Comparison between linear and spiral DDA MS imaging.
(a) and (b) illustrate the step motion and size, respectively, while (c)-(h) demonstrate image quality obtained from both with high mass accuracy. Reprinted with permission from (OuYang, Chen & Li, 2015).
Singly-charged ions produce the simplest spectra, which means that MALDI-TOF analyses allow for the widest mass range that can be analyzed. Tandem TOF (TOF/TOF) mass analyzers have a theoretically infinite mass range, with analytes larger than 50 kDa being imaged with high signal (Leinweber et al., 2009; van Remoortere et al., 2010). Unfortunately, most TOF/TOF mass spectrometers lack the mass accuracy and resolution that allow for differentiation between closely massed neuropeptides of interest (Verhaert et al., 2010)). On the other hand, Orbitraps and FTICR instruments have a limited mass range, leading to several larger neuropeptides of interest not being imageable as singly-charged ions. Several methods to handle these larger mass analytes have been developed, such as in situ digestion and generating multiply-charged ions by MALDI (Cillero-Pastor & Heeren, 2014; Dreisewerd, 2014; Groseclose et al., 2007). Dependent on the sample preparation conditions, multiply charged ions are usually produced by using laserspray ionization (LSI) at atmospheric, intermediate, or high vacuum (Chen, Lietz & Li, 2014; Hale et al., 2021; Inutan, Wang & Trimpin, 2011; Trimpin et al., 2011). Trimpin and coworkers have analyzed a 12+ charge state cytochrome c by atmospheric pressure (AP) - MALDI on a Thermo Fisher Q Exactive mass spectrometer (Trimpin et al., 2010).
Data processing represents challenges as well, particularly in high-throughput data collection. Pipelines for automated identification of unique peptides were developed (e.g. MSI-Query) (Bruand et al., 2011). Software packages developed to view MS images are vendor-specific (e.g. Thermo Fisher ImageQuest and TissueView, Waters HDImaging, Bruker SCiLS lab etc.) or more generic/open source (e.g., MSiReader, Cardinal, msIQuant) (Bemis et al., 2015; Källback et al., 2016; Robichaud et al., 2013). Many packages are utilized to identify masses unique to the tissue (which may be known or unknown), such as the program written to perform accurate mass matching with an intensity threshold (Buchberger et al., 2020b). A particular program has been published for normalization and quantitation-based studies (Kallback et al., 2012). This software enables the quantitation of SP in mouse brain structures, which correlated well with previous studies (Kallback et al., 2012).
One area that has gained a lot of attention is spatial resolution, defined as how small the pixels can get in MSI. Higher spatial resolution allows MSI of smaller biological tissues down to even single cells (Boggio et al., 2011; Xie & Fidler, 1998; Zimmerman, Rubakhin & Sweedler, 2011). Two factors play major roles in the maximum resolution achievable in MALDI MS imaging: matrix crystal size and focusing of the laser. The crystal size is dependent on the matrix and application method used (see Sample Considerations). It is especially instrumental advancements which have provided most improvements in this context. Commercial instruments allow for small pixels with oversampling, but this can lead to poor signal in tissues with already low analyte concentrations. However, with home-built instruments, some groups have achieved 5-micrometer resolution without oversampling (Guenther et al., 2011; Kompauer, Heiles & Spengler, 2017; Mark, Maasz & Pirger, 2012; Rompp & Spengler, 2013), which allows imaging discrete cellular structures (Boggio et al., 2011; Dueñas, Essner & Lee, 2017; Xie & Fidler, 1998; Zimmerman, Rubakhin & Sweedler, 2011). It should be noted that this typically lowers the throughput of the instrument due to the longer acquisition time, but some companies have developed scanning laser beams to lessen this time (Ogrinc Potocnik et al., 2015). Alternatively, Zimmerman et al. have achieved this by placing individually stretched cells on an ITO-coated slide and analyzing them with a Bruker Ultraflex II MALDI-TOF/TOF (laser beam diameter is between 5 to 30 μm), which allowed them to acquire MS and MS/MS images of neuropeptides throughout the cell body (>0.5 mm diameter) (Zimmerman et al., 2009). The Bruker rapifleX MALDI TOF/TOF mass spectrometer has become a popular choice for peptidomics (Vu, DeLaney & Li, 2020) due to its high spatial resolution (<20 μm) and the fast laser repetition rate, and its ability to scan the full area of a pixel while the sample stage moves continuously, allowing rapid acquisition of MSI data. Continued instrumental development allows researchers to do more single cell studies, and quality reviews discuss the next challenges that need to be met in order to advance the field (Berman, Fortson & Kulp, 2010; Boggio et al., 2011; Xie & Fidler, 1998; Zimmerman, Rubakhin & Sweedler, 2011).
B. Sample Considerations
Proper sample handling is crucial for maintaining the spatial distribution and abundance of biomolecules in a sample, allowing for maximum spatial resolution, sensitivity, and reproducibility of an MSI experiment (Goodwin, 2012). Studies of post-mortem changes in peptide and protein abundance in brain tissue demonstrate the necessity for sample collection protocols that limit sample degradation (Goodwin et al., 2008; Skold et al., 2007). To preserve sample integrity, samples are typically flash frozen either using liquid nitrogen or dry ice. Alternatively heat stabilization, often using e.g. a Stabilizor T1, (Goodwin et al., 2010; Sturm et al., 2013b). Fixation methods, such as formaldehyde-fixed paraffin embedding (FFPE), are commonly used to preserve samples in biomedical research, but this procedure is reported not be compatible with MS imaging FFPE is known to result in crosslinking between peptides and proteins, which is predicted to have a negative impact on MSI. Optimized protocols for MSI on FFPE material involve deparaffination, antigen retrieval, and trypsin digestion before analysis (Casadonte & Caprioli, 2011; De Sio et al., 2015). MS imaging of proteins and neuropeptides has been performed in rat brain FFPE samples, after deparaffination and tryptic digestion, and in Penaeus monodon shrimp (Chansela et al., 2012; Lemaire et al., 2007; Stauber et al., 2008). A protocol for performing MS imaging of neuropeptides from FFPE tissue without antigen retrieval and enzymatic digestion has also recently been developed (Paine et al., 2018). Alternatively, alcohol fixation methods have been used to fix samples without any of the complications of FFPE (Chaurand et al., 2008). The PAXgene system is an alcohol fixation system commercially available that can be used to fix samples prior to MS imaging, although use of this system is not as widespread as FFPE (Ergin et al., 2010). Interestingly, DHB matrix can also be used as a one-step tissue preservation and peptide extraction solvent (Alim et al., 2019; Romanova, Rubakhin & Sweedler, 2008). Multiple reviews discuss sample preparation in more depth (Buchberger et al., 2018; DeLaney, Phetsanthad & Li, 2020; Gemperline, Chen & Li, 2014; Goodwin, 2012; OuYang, Liang & Li, 2015).
Traditionally, prior to MSI analysis, the typical (frozen) tissue samples are sectioned into 10-20 μm thick slices, which is roughly the thickness of a single mammalian cell (Crossman et al., 2006). Before sectioning, samples are usually embedded in a support substance to aid in sectioning. These can be water and gelatin that do not interfere with the MSI analyses of neuropeptides. Other polymer-based support substances such as optimal cutting temperature (OCT) medium is known to contaminate the sample and suppress ion formation (Buchberger et al., 2018; OuYang, Liang & Li, 2015). A novel embedding material, poly[N-(2-hydroxypropyl)methacrylamide] (pHPMA), was tested on mouse lung and bumblebee samples and it was found to be suitable for MALDI MS imaging with low background signal and ion suppression effects (Strohalm et al., 2011). Also egg yolk seems to be an appropriate embedding material for MSI of neuropeptides in rat pituitary, preventing OCT contamination (Sosnowski et al., 2015). The sectioned samples are transferred to a simple glass or a metal coated glass slide, depending on instrumentation, using either a thaw-mount method or with double-sided tape (Goodwin et al., 2012). Certain intact tissues are thin enough to bypass the sectioning step and can be directly analyzed by MALDI MS. Examples of this are crustacean pericardial organs and cardiac ganglion (Buchberger et al., 2020c; DeLaney & Li, 2020; Zhang et al., 2018c), and the insect corpus cardiacum (Verhaert et al., 2007; Verhaert et al., 2010). It is important to note that fragile samples, such as pericardial organs, benefit from immediate MS analysis after dissection to prevent tissue degradation.
There are multiple treatment steps that can be taken before matrix application. For protein analysis, washing tissue sections with organic solvents such as xylene, chloroform, or alcohols has been shown to increase detection through removal of contaminating compounds, such as lipids and salts (Lemaire et al., 2006; Seeley et al., 2008). However, washing steps risk to cause delocalization or loss of low molecular weight or hydrophilic neuropeptides if not optimized (OuYang, Liang & Li, 2015; Yu et al., 2014). Nonetheless, proper optimization of tissue washes has been shown to be effective at enhancing neuropeptide signal (Vu, DeLaney & Li, 2020). Reduction of salt adducts has also been shown using a condensation/matrix recrystallization procedure after matrix deposition (Monroe et al., 2007). An aqueous MS imaging tissue wash containing sodium phosphate salts resulted in detection of a complementary cohort of neuropeptides compared to control, unwashed tissue (Vu et al., 2021). MSI of neuropeptides in Aplysia nervous tissue utilized a tissue stretching method to fragment the tissue into small pieces, which minimizes analyte diffusion and salt adduct formation (Zimmerman et al., 2009). Another option is the application of trypsin to the sample to digest larger proteins or certain large neuropeptides to the mass range ideal for higher resolution instrumentation (Cillero-Pastor & Heeren, 2014; Groseclose et al., 2007). Optimization of digestion times, proteases, and matrix application has been performed in brain tissue to improve the repeatability of trypsin digestion (Diehl et al., 2015; Heijs et al., 2015). Other protocols look to improve trypsin digestion reliability and reproducibility utilizing graphene oxide-immobilized enzyme reaction (Jiao et al., 2013) or microwave irradiation and hydrogel discs (Taverna, Norris & Caprioli, 2015).
The choice of matrix and matrix application method is critical for ionization of the target analytes while limiting diffusion in MALDI MS analysis (Kaletaş et al., 2009). Common matrices include CHCA and DHB for the analysis of peptides and sinapinic acid and DHB for the analysis of larger proteins. Matrix concentration, solvent composition, and deposition temperature are factors that impact matrix crystal size and therefore spatial resolution. Hulme et al. observed that a higher concentration of matrix and higher deposition temperature (i.e. drier deposition) resulted in high spatial resolution (15-25 μm), but using a lower concentration and temperature resulted in more neuropeptide identifications but at lower spatial resolution (50 μm) (Hulme et al., 2020). Many new matrices are being investigated for application in MS imaging experiments of all analyte types, from metabolites up to proteins (Buchberger et al., 2018; Dreisewerd, 2014).The derivatization of chemical compounds with amines, including catecholamine neurotransmitters and neuropeptides, by reacting primary amines with pyrylium salts have been proposed as a novel matrix for MSI of primary amine compounds, which are usually challenging to detect (Shariatgorji et al., 2015). Additionally, graphene has been used as matrix on brain tissue to detect lipids and small peptides (Friesen et al., 2015). The choice of matrix is important for studies using LSI to produce multiply-charged ions. For example, when using 2-nitrophologlucinol as a matrix, multiply-charged ions are produced at both vacuum and atmospheric conditions, but some matrices, like CHCA, only produce singly charged ions at all conditions (Chen et al., 2018; Inutan et al., 2011; Inutan et al., 2012). To apply matrix for MS imaging experiments, several different options are available, including robotic spotters, airbrush, and automated spraying devices. Robotic spotters, such as acoustic droplet ejectors (Aerni, Cornett & Caprioli, 2006) and inject printers (Baluya, Garrett & Yost, 2007; Franck et al., 2009) apply small amounts of matrix in spots of approximately 150-200 um diameter in an ordered array across the tissue. Automated spraying systems, such as the TM-Sprayer or M5 Sprayer (HTX Imaging, NC, US), pneumatic spraying devices, and the ImagePrep (Bruker Daltonics, Bremen, Germany) vibrational sprayer, allow more control compared to manual airbrush. Use of these automated methods has been shown to produce higher quality data compared to other methods (Gemperline, Rawson & Li, 2014). Our group has reviewed recent advances (from years 2017-2020) in MS imaging washes, matrices, and other sample preparation considerations for neuropeptide analysis (Buchberger et al., 2018; Vu, DeLaney & Li, 2020).
C. Special Applications of MS Imaging
While the application of MSI to directly answer biological questions has boomed, MSI has also been utilized for new applications. For example, the 3D analysis of structures in a heterogeneous tissue (Chen et al., 2009a; Dueñas, Essner & Lee, 2017; Jones et al., 2012; Trede et al., 2012). For example, consecutive sections have been analyzed in 3D to demonstrate the spatial variability of several crustacean neuropeptides (Chen et al., 2009a). MSI has been used in 3D cell culture studies (Fernandes, 2004; Li & Hummon, 2011), e.g. to understand the depth of drug penetration or the production of different metabolite due to changing environments (e.g. normoxia vs. hypoxia). Another analytical technique that has been combined with MSI is microfluidics, to study neuropeptide secretion from a cell (Jo et al., 2007; Zhong et al., 2012). Finally, unlike spot analysis, analyte traces analyzed by MSI has become a way to add temporal information to this spatial technique (DeLaney & Li, 2019a; Wang et al., 2011; Zhang, Jia & Li, 2012; Zhang, Kuang & Li, 2013; Zhang et al., 2012b). Initially, this was used to analyze CE or LC traces of tissue extracts, but MSI of traces has evolved to direct analysis of microdialysates (OuYang, Liang & Li, 2015). This combination has also proven to be remarkably accurate for quantitative analysis, allowing both relative and absolute concentrations of neuropeptides to be obtained (Zhang, Kuang & Li, 2013; Zhang et al., 2012b).
Advances in Quantitation
To analyze a system for biological relevance, quantitative tools are necessary. Most techniques are compatible with both ESI and MALDI sources, but special considerations should be made for either ionization method. ESI is well known for consistency, but often ESI is done in conjunction with LC separation. Any run-to-run variation will need to be corrected for by bioinformatic tools. MALDI, on the other hand, is notorious for inconsistent ionization (such as due to variability in matrix crystallization). This makes MALDI, without further methodological developments, inherently semi-quantitative. With that in mind, we will focus below on the development of relative (i.e., comparative) vs. absolute (i.e. actual) quantitation with both label vs. label-free methodology for the study of neuropeptides (Figure 6). Many recent quality reviews exist, and only the major contributions will be highlighted below (Buchberger, Yu & Li, 2015; Fricker, 2018; Fricker et al., 2006; Li & Sweedler, 2008; Romanova, Dowd & Sweedler, 2013; Yin et al., 2011).
Figure 6. Two major types of quantitation in MS.
(A) Label-free quantitation strategies, include intensity comparisons and spectral counting. (B) Label-based quantitation. These techniques can be done at both MS1 and MS2 levels.
A. Labeling-Based Methods
The incorporation of standard isotopes has revolutionized MS for quantitative analysis. The more variations of isotopes we have, the more samples we will be able to compare, which will thus increase analysis throughput. There are several different ways to incorporate isotopes into an analyte of interest, including in vivo metabolic labeling. This is done usually by culturing cells with a heavy isotope, for example in the form of an amino acid, allowing it to be incorporated during the synthesis of other cells (Ong et al., 2002; Potts et al., 2016). Heavy amino acids have also been added into the diets of animals (Kruger et al., 2008; Zanivan, Krueger & Mann, 2011) and plants (Lewandowska et al., 2013). A simplified protocol has been created to quantify fruit fly neuropeptides by growing differential isotopically labeled yeast that can be fed to different groups of flies (Kunz et al., 2018). Isotopic neuropeptides can also be administered intranasally or intravenously into animals (Lee et al., 2018). While decreasing variability in the population analyzed, full incorporation of the isotopes can take a long time depending on cell turn over, especially in animals, leading to a high cost. The number of samples that can be compared is limited by the number of isotopes of an element. This methodology has been mainly used in protein quantification (Geiger et al., 2010; Kruger et al., 2008; Lewandowska et al., 2013; Ong et al., 2002; Potts et al., 2016; Zanivan, Krueger & Mann, 2011), although it could be adapted for neurochemical cell culture studies.
Another variation of this MS labeling method is in vitro chemical tags. Generally, the analytes of interest are derivatized with a chemical tag that includes stable isotopes, which produce well-defined mass differences between the samples at either the MS1 or MS2 level. For neuropeptidomic studies, MS1-based quantitation is becoming more and more common. In particular, duplex dimethyl labeling has been used thanks to its simplistic and quick labeling on all primary amines (e.g., N-terminus and ε-lysine). For example, the Li lab has utilized this method for studying the dynamic changes in neuropeptides due to environmental stress (Buchberger et al., 2020a; Buchberger et al., 2020b; Chen et al., 2010b; Chen et al., 2014; Liu et al., 2019; Zhang et al., 2015). Wilson et al. achieved in vivo quantitation of Leu-enkephalin and Met-enkephalin after on-column dimethyl labeling of microdialysis perfusate from rat brain (Wilson, Jaquins-Gerstl & Weber, 2018). Dimethyl labeling was expanded from 2 to 5 plex (Boersema et al., 2008; Buchberger et al., 2020b; Hsu, Huang & Chen, 2006; Tashima & Fricker, 2018). Isotopic dimethyl N,N-leucine (iDiLeu) and mass defect-based N,N-dimethyl leucine (mdDiLeu) also contains five spaced channels, which allows for relative or absolute quantitation (Greer et al., 2015; Zhong, Frost & Li, 2019). By labeling 4 channels with neuropeptide standards to construct a calibration curve, the fifth channel can be used to calculate the absolute concentration of an unknown sample. Care should be taken that, when all 5 channels are in use and ESI is chosen as the ionization technique, isotopic impurities and charge state differences may lead to overlapping peaks and thus inaccurate quantitation (Greer et al., 2015). In terms of MS1-based quantitation, other options exist to label the N-terminus, such as succinic anhydride (Bark, Lu & Hook, 2009; Fricker, 2006; Hou, Xie & Sweedler, 2012; Rubakhin & Sweedler, 2008), acetic anhydride (Che & Fricker, 2002), and 4-trimethylammoniumbutyryl (Che, Biswas & Fricker, 2005). Amino acid specific labels, including isotopic-coded affinity tag (ICAT), metal-coded affinity tag (MeCAT), and tyrosine-specific cysteine labeling (Ahrends et al., 2007; Choi, Pennington & Wood, 2010), are also accessible to study neuropeptides. All these labeling schemes come with similar considerations to dimethyl labeling, iDiLeu, and mdDiLeu. While a balance between multiplexing and spectral complexity is a major concern, the development of tags with smaller spaces alleviates some of this burden. Unfortunately, these small spacing usually requires high-resolution instrumentation, which may not be readily available for many labs. For example, neutron-encoded (Hebert et al., 2013b), mdDiLeu (Hao et al., 2017), and dimethyl pyrimidinyl ornithine (DiPyro) (Frost, Buchberger & Li, 2017) tags all take advantage of isotopic mass defect, but high multiplexing requires resolution only achievable by top tier instrumentation.
The use of chemical tags that quantitate at the MS/MS level can also decrease the MS1 spectral complexity occurring above. Instead of producing mass shifts at the MS1 level, all the differentially labeled peptides occur at the same mass in the initial MS1 scan. If the associated peak is selected for fragmentation, characteristic reporter ions are created, usually in the low mass range where no interference occurs. Unlike at MS1 level, where theoretically every analyte can be quantified, one is limited by the peptides which get selected for fragmentation. Thus the duty cycle of an instrument can play a major role in the depth of quantitation achieved. Many commercial tags, such as isobaric tags for relative and absolute quantitation (iTRAQ) and tandem mass tags (TMT), have been used (McAlister et al., 2012; Rubakhin & Sweedler, 2008). iTRAQ has even been used for single cell analysis, allowing the relative quantities of peptides to be obtained by MALDI MS (Rubakhin & Sweedler, 2008). Unfortunately, the cost of these commercially available labels limits their use. N, N-dimethyl leucine (DiLeu) is one, low cost example for MS2 level quantitation, and it has been expanded from 4-plex to 21-plex quantitation (Frost, Feng & Li, 2020; Frost, Greer & Li, 2015; Frost et al., 2015; Liu et al., 2020; Xiang et al., 2010). Recently, DiLeu and iDiLeu have been combined to form a strategy called hybrid offset-triggered multiplex absolute quantification (HOTMAQ) which enables the formation of an internal standard curve at the MS1 level, peptide sequencing and identification at the MS2 level, and peptide quantification at the MS3 level (Zhong et al., 2019). With the multitude of channels all these tags contribute, both absolute and relative quantitation is possible. Most of these tags have not been applied to neuropeptide quantitation, but they would provide a practical way to compare several samples in one instrumental run.
B. Label-Free Methodology
Label-free quantitation techniques are more frequently used in the study of neuropeptides. Unlike labeling strategies, label-free methods allow one to compare an infinite number of samples. The simplest label-free method is based on signal intensity, meaning that the signal intensity in the spectra, or more accurately the area under the curve in the LC chromatogram, correlates with the analyte concentration. Relative quantities are easily found by just comparing samples at either the MS1 or MS2 (i.e., MS/MS) level, although peak alignment and other post-processing aspects need to be considered due to run-to-run variability (Jiang et al., 2012; Johansson et al., 2006; Ranc et al., 2012). For example, Ranc et al. utilized the extracted ion chromatograms to obtain relative quantities of different endogenous peptides in the tree shrew visual system (Ranc et al., 2012). It should be noted that at the MS/MS level, multiple reaction monitoring, or monitoring of only specific, characteristic fragments, and parallel reaction monitoring, or monitoring of all fragments, can lead to lowering the limit of detection by 100-fold (Bobba, Resch & Gutheil, 2012; DeAtley et al., 2018; Pailleux & Beaudry, 2014; Saidi, Kamali & Beaudry, 2019; Song & Liu, 2008; Wang, Chung-Davidson & Li, 2014; Yang et al., 2017).
In order to acquire the absolute concentration of neuropeptides, (a) a calibration curve is required (Chung-Davidson et al., 2020; Schmerberg, Liang & Li, 2015; Song & Liu, 2008; Wang, Chung-Davidson & Li, 2014); (b) a synthetic, isotopic internal standard, also known as an AQUA peptide is added (Bozzacco et al., 2011; Ozalp et al., 2018; Salem, Nkambeu & Beaudry, 2018); or (c) a peptide standard similar to the peptide of interest (Dong et al., 2018) to be used as a proxy. Several software packages assist in processing these large datasets, including commercial software packages (SIEVE, PEAKS, Proteome Discoverer…) or open access platform (e.g., Skyline). Several groups have developed their own pipelines, such as using accurate mass time (AMT) (Wu et al., 2015a), informed quantitation (IQ) (Wu et al., 2015a), and DeCyder MS (Johansson et al., 2006; Kaplan et al., 2007).
Many of the above informatics tools also assist in another label-free quantitation (LFQ) method: spectral counting. Unlike peak area/signal intensity measurements, spectral counting is dependent upon the number of times an analyte is selected for fragmentation. Similar to MS/MS label-based quantitation, only high concentration molecules are analyzed due to the limited duty cycle of an instrument. Also, care should be taken on instrumental parameters, such as scan and exclusion parameters in order to increase sampling depth (Zhou, Liotta & Petricoin, 2012). Relative comparisons are easily done by comparing the number of MS/MS spectra collected between analytes, although validation of the smaller differences will be particularly important. Furthermore, only estimates can be made based upon total protein concentration (Neilson et al., 2011). When comparing SIEVE peak area analysis and spectral counting on peptides in the rat suprachiasmatic nucleus, it was revealed that spectral counting provided a richer characterization of differences in differential peptide abundance when rats were analyzed at different circadian rhythm points (Southey et al., 2014). Furthermore, when compared to SILAC and spectral counting for quantifying proteins, spectral counting was found to be able to quantitate ~50% more proteins prior to a limit being set (Collier et al., 2010). This shows the power of spectral counting and label-free quantitation for neuropeptidomic analysis, and it is likely that these approaches will be increasingly used.
C. DIA Quantitation
DIA strategies can be applied to improve quantitative studies, however the use of label-based quantitation adds additional complexity to the MS or MS/MS spectra collected. This makes spectral deconvolution and accurate quantitation difficult to achieve. Therefore, LFQ is commonly used in conjunction with DIA. In fact, a recent comparison study between LFQ DIA and isobaric tag labeling with DDA demonstrated similar performance between workflows (Muntel et al., 2019). Performing 10 LFQ DIA analyses demonstrated better quantitative accuracy while a single multiplex TMT labeled DDA analysis resulted in an increase in identified proteins and quantitative precision (Muntel et al., 2019). This demonstrates the capability of LFQ DIA for quantitation; although, to fully leverage the benefits of DIA, specific DIA labeling strategies need to be developed. mdDiLeu (Zhong et al., 2020), NeuCode SILAC (Hebert et al., 2013a; Hebert et al., 2013b), and MdFDIA (Di et al., 2017), are labeling strategies that have been successful for DIA quantitation. The latter two rely on metabolic labeling and are thus restricted to cell culture applications, and none of these techniques have been applied to neuropeptidomics. Developments in the DIA quantitative analysis to more broadly study the brain proteome are summarized by Li et al. (Li et al., 2020a).
Parker and colleagues applied a LFQ DIA strategy in a targeted phosphoproteomics analysis to understand signaling of the peptide hormone insulin (Parker et al., 2015). The increased throughput and reproducibility, enabled by DIA analysis, led to the quantitation of 86 protein targets affected by insulin (Parker et al., 2015). In a new Skyline software application (MacLean et al., 2010), Schmerberg et al. performed quantitation of LFQ DIA MS/MS data in a pseudo-multiple reaction monitoring analysis of crustacean neuropeptides (Schmerberg, Liang & Li, 2015). They were able to identify and quantify several neuropeptides from microdialysate and their changes across the feeding study illustrating the sensitivity of the method (Schmerberg, Liang & Li, 2015). Saidi et al. evaluated the utility of label-free and isotopic dilution DIA methods for targeted quantitation of neuropeptides and found an increase in variance when compared to PRM methods (Saidi, Kamali & Beaudry, 2019). This could be attributed to the increase in cycle time for the DIA method, which decreases the points per chromatographic peak acquired. DeLaney and Li optimized the DIA duty cycle for crustacean neuropeptides by considering various isolation windows and m/z ranges. They also evaluated the quantitative accuracy and observed experimental errors between 18.0% and 32.8% (DeLaney & Li, 2019b). Potential improvements to this method could include the use of label-based quantitation. While the capabilities and applications for DIA has expanded over the years, it will benefit from additional improvements and new labeling strategies for accurate and reproducible quantitation.
D. Special Considerations: MS Imaging
High throughput data collection is key in developing new analytical techniques. Thus, the application of quantitative methods to imaging was a natural transition to acquire both spatial and quantitative information in a single instrumental run. Some applications have been discussed briefly above (see MS Imaging: Special Applications) (Zhang, Kuang & Li, 2013; Zhang et al., 2012b; Zhong et al., 2012), but the streamlining of methods has obtained a lot of attention recently for drugs and metabolites (Pirman, 2015; Sun & Walch, 2013). While this has not been fully developed for neuropeptidomics, it could be easily implemented in the future. It should be noted that these techniques still require further development to become more common practice in the scientific community (Cillero-Pastor & Heeren, 2014).
As stated above, there are label-free and label-based techniques for acquiring quantitative information from samples. LFQ is the most commonly used in MSI, including the use of a calibration curve or an internal standard (Clemis et al., 2012; Goodwin et al., 2012; Groseclose & Castellino, 2013; Hamm et al., 2012; Lanekoff & Laskin, 2017; Nakanishi, Nirasawa & Takubo, 2014; Rodrigues et al., 2014; Shariatgorji et al., 2014). In these cases, usually the standard(s) are either spotted onto the tissue or added to the MALDI matrix solution prior to its application. Alternatively, the use of multiple isotopically labeled standards can be sprayed onto the tissue section for use as internal standards (Dewez et al., 2021). Koeniger and others have taken a more unique approach by taking nearby, separate sections for MS imaging and LC-MS quantitation, since serial sections have similar analyte concentrations (Koeniger et al., 2011). This approach requires homogenous tissues. MSI, label-based quantitation applications are still novel. The only published example utilizes a duplex-isotopic immunohistochemical staining azo dye which, after laser energy absorption, produces signature reporter ions separated by 5 Da (Wang et al., 2015a). While all these methods seem promising, without the ability to process this data quickly, the throughput of quantitative MSI is limited. Some groups have produced software for on-tissue calibration curve quantitation, both open source (e.g. MSiReader) or commercial (e.g. SCiLS or Quantinetix) (Kallback et al., 2012; Robichaud et al., 2013). More effort needs to be applied to developing additional bioinformatics tools in this area.
The generation of a calibration curve using a peptide standard to absolutely quantify neuropeptides is considered the gold standard. This can be performed on a variety of instruments and does not require many biological replicates to produce confident results. However, peptide standard synthesis is expensive. Additionally, if the neuropeptides selected for quantification exclude other co-modulating neuropeptides involved in a particular biochemical pathway, incorrect conclusions might be drawn. As only a small number of discovered neuropeptides have been functionally evaluated, there is high risk of not selecting all the neuropeptides involved in the pathway. In this case, it is better to perform global profiling analyses to detecting as many neuropeptides as possible simultaneously. Therefore, the next best alternative for quantitation is to either incorporate isotopes into animals before sample collection or to chemically derivatize the animal samples after collection. For both methods, the optimal means would involve detection of the same neuropeptides in all conditions, while also not detecting any non-modified neuropeptides. Additionally, in chemical derivatization, a 100% labeling efficiency would be achieved. Overall, global profiling of changes in neuropeptide expression can serve as a foundation to understand neuropeptide function and dysregulation.
Functional Studies
Neuropeptides impact a large and diverse array of physiological processes (Insel, 2010; Mills et al., 2020; Neumann & Landgraf, 2012; Steinhoff et al., 2014; Wang, Xu & Kang, 2021; Xu et al., 2020). Functional elucidation is not trivial notably due to neuropeptide co-transmission capabilities (Nusbaum, Blitz & Marder, 2017) and pleiotropic nature (Souza-Moreira et al., 2011). Changes in neuropeptide abundance and localization can act as a foundation for functional studies since dysregulation of these characteristics indicate an abnormal or disease state (DeLaney, Buchberger & Li, 2018) (see elsewhere in this review). Thus, MS-based quantification of neuropeptides can be exploited to understand neuropeptide expression level changes under physiological and pathological conditions. For example, Ye et al. profiled neuropeptide expression changes due to differential food intake and functionally validated the role of significantly changed neuropeptides by injecting them into rats (Ye et al., 2017). This section focuses on physiology- and microdialysis-based functional studies where neuropeptides are the target analyte. Yet, it is also worth noting that neuropeptide receptor dynamics also play a critical role in neuropeptide function (DeLaney et al., 2018).
A. Physiology-based Functional Studies
Besides MS, other techniques are often used to understand neuropeptide function, and these characterizations are critical for development of therapeutics. For example, pituitary adenylate cyclase activating polypeptide (PACAP), known to improve cornea health, is shown by Kovacs et al. to be resistant to degradation in solution, demonstrating its potential for use in eye drops (Kovacs et al., 2020). A common technique to investigate neuropeptide function is overexpression of the peptide in an animal model. Transgenic mice overexpressing thyrotropin-releasing hormone exhibit higher blood pressure and heartbeat rate (Landa et al., 2020). Since the development of transgenic animal models is difficult and costly, so alternative methods are often preferred.
Neuropeptide function are often characterized by examining physiological effects in vitro or ex vivo. Such studies allow the researcher to control experimental parameters better than in in vivo experiments. For example, somatostatin/allatostatin-C ArSS2 standards have a relaxation effect on dissected starfish tube foot, apical muscle, and cardiac stomach muscle contractions (Zhang et al., 2020b). Muscle contractions are typically recorded using a timer or by connecting the tissue to a force-displacement transducer or similar instrument recording contraction force. Manual counting is advantageous when studying small animals as employed in a recent study on the effect of adipokinetic hormone Carmo-HrTH-II neuropeptide on heartbeat rate (Katali, Marco & Gade, 2020). Since certain invertebrates, including decapod crustaceans, have neurogenic hearts, neuropeptide modulation of cardiac function has become a field of interest, broadly reviewed by Calabrese, Norris, & Wenning (2016) (Calabrese, Norris & Wenning, 2016). Marciniak et al. showed that FMRF6 causes a decrease in beetle heartbeat rate and an increase in hindgut contractions (Marciniak et al., 2020). Dickinson et al. perfused shrimp pyrokinin PevPK2 neuropeptide onto a lobster heart to observe an increase in heartbeat rate and amplitude and decrease in heart contraction duration. Altering the peptide sequence resulted in a loss of activity (Dickinson et al., 2015). The Dickinson group used semi-intact heart preparations to evaluate the neuropeptide modulation of heart (Wiwatpanit, Powers & Dickinson, 2012) and also published a review on crustacean neuropeptide modulation of pattern generating systems (Dickinson, Qu & Stanhope, 2016). Cardiac assays have also been performed on mammals. Studneva et al., administered forms of galanin to myocardial injury-induced rats and recorded blood pressure and heart rate in vivo (Studneva et al., 2019).
Additional methods to investigate neuropeptide function measure biochemical effects. Wei et al. cultured crab hepatopancreas tissue and applied crustacean cardioactive peptide (CCAP), measuring an increase in nitric oxide and resulting improved bacterial clearance in the medium (Wei et al., 2020). To investigate the impact of neuropeptides on reproduction, Chieu et al. incubated dissected sea cucumber ovarian tubules in solutions containing gonad-stimulating peptide and observed oocyte maturation (Chieu et al., 2019b). Hao et al. injected newly synthesized diapause hormone (DH)-like peptides into locusts and found some peptides to induce diapause in eggs (Hao et al., 2019). Atkins et al. applied neuropeptides, including arginine vasopressin to excite rat optic nerves ex vivo to evaluate their involvement in the regulation of circadian rhythm (Atkins et al., 2018).
One strategy is to minimize sample handling, as seen in the use of microfluidic platform to culture neurons in a capillary and directly analyze secreted neuropeptides by MS (Lee et al., 2016). While in vitro and ex vivo experiments have their clear benefits, there is a push towards in vivo approaches, particularly when translation to therapeutics is aimed for.
B. Microdialysis-based Functional Studies
Microdialysis probes in or adjacent to the location injected with peptides can be used to collect local perfusates. Guvenc-Bayram et al. observed an increase in prostaglandin in mice hypothalamic injected with nefastin 1, indicating that this peptide activates the arachidonic acid-cyclooxygenase and -lipoxygenase signaling pathway (Guvenc-Bayram et al., 2020). Our lab has recently investigated neuropeptides implicated in circadian rhythm using microdialysis (Liang, Schmerberg & Li, 2015), after the development of a protocol for the in vitro microdialysis of a neuropeptide standard as well as the in vivo microdialysis sampling of neuropeptides from a live crab (Behrens & Li, 2010). Using microdialysis coupled with MS, Mabrouk et al. measured an increase in oxytocin and arg-vasopressin levels in rat brain (Mabrouk & Kennedy, 2012).
Bulbul et al. administered neuropeptide-S into Parkinson’s disease-induced rats and observed increased dopamine levels (collected via microdialysis) 7 days after the administration. This suggests that the peptide has protective effects in the brain (Bulbul et al., 2019). Grund et al. examined potential anxiety disorder therapeutics and saw that neuropeptide S stimulates oxytocin release (Figure 4) (Grund et al., 2017). Cui and Smith studied the neuronal regulation of obesity and demonstrated an increase in agouti-related peptide release when Gs-linked G protein-coupled receptors were activated (Cui & Smith, 2019). Willie and collaborators combined intracerebral microdialysis and electroencephalography/electromyography with motor activity monitoring to study the effect of orexin neuropeptides in brain injury (Willie et al., 2012). In addition to roles in biochemical signaling, certain neuropeptides have measurable behavioral effects. Lee et al. delivered oxytocin neuropeptide into mice and observed a decrease in the rate by which mice self-administered the drug methylphenidate (CNS stimulant) along with differential regulation of dopamine levels (collected via microdialysis from mice that were randomly implanted in the right or left brain side) between different brain regions (Lee et al., 2019). A logical next step is to expand the number of simultaneously measurable characteristics, particularly during in vivo experiments, and to increase the sensitivity for neuropeptide detection, such as by improving sample preparation methods (see Sample Preparation section).
Figure 4. In hypothalamic slices, neuropeptide S (NPS) stimulates silent oxytocin (OXT) neurons via NPS receptor (NPSR) but does not stimulate active OXT neurons.
(A) Schematic drawing of the PVN OXTpr-GCaMP6s virus infusion and subsequent [Ca2+] imaging of OXT neurons. (B) Basal activity of two distinct subpopulations of OXT neurons (dark gray: active; light gray: silent) illustrated by typical ΔF/F0 traces. Pie charts represent the proportion of active (up) and silent (down) OXT neurons: n slices (ns) = 11, n OXT neurons (nn) = 237. (C) Pie charts of proportion of responsive OXT neurons to NPS application alone (2 μm, 20 s; ns = 11, nn = 24 of 237; green) or in the presence of NPSR antagonist (SHA-68 100 μm, >15 min; ns = 6, nn = 3 of 135; light blue) and typical ΔF/F0 traces. Pseudo-color video extract of identified OXT neurons through GCaMP6s imaging [Ca2+] in control conditions (gray), in presence of NPS (green) or NPS + SHA-68 (light blue) (stacks of 50 images/10 s of recording). Scale bar, 20 μm. (D) Relative AUC increase and maximal ΔF/F0 of OXT neurons in presence of NPS (ns = 11; green) or NPS + SHA-68 (ns = 6, light blue). Only response duration of OXT neurons in presence of NPS (ns = 11; green) are represented here. White circles represent average value per slice. *p < 0.05 (Student's t test). **p < 0.01 (Student's t test). Reprinted with permission from (Grund et al., 2017).
Conclusions
In the last decade, significant advances in mass spectrometry instrumentation and associated technologies have accelerated the progress of neuropeptide research, enabling high throughput neuropeptidome characterization. As the biological importance of neuropeptides is increasingly realized, we predict that more people will be attracted to study them and conduct more in-depth investigations. However, compared to well-established proteomic workflows and tools, there are still many technological gaps to be filled, and implementation of advancements in proteomics tools should be more readily applied to neuropeptidomics. Techniques capable of reliably enriching scarcely distributed neuropeptides and removing interfering substances are in high demand. As neuropeptides vary in length and structure, there is a need for customized MS approaches to be developed based on each particular family, class, and even isoform of neuropeptides being targeted to enable obtaining comprehensive MS/MS spectra. For MSI, areas of interest are better robust sample preparation techniques, improving spatial resolution, increasing throughput, and development of quantitation methods. Algorithms that are able of integrating prohormone cleavage preferences would be beneficial in performing mature neuropeptide prediction from genomes as they are increasingly being sequenced. Further advances in bioinformatics must keep up such that all MS data will be interpreted in a convenient fashion while providing rich chemical information. Although empirical determination of individual neuropeptide functions is highly valuable, the time it takes to do so can be considered a bottleneck step in the overall pipeline from discovery to therapeutics. To improve and facilitate interpretation of neuropeptidomics data, methods capable of elucidating neuropeptide co-modulation must be developed. Finally, further development of sensitive, reliable quantitation approaches that can handle limited sample amount will be key to allow cross comparisons of neuropeptides in a high throughput manner. While the field of neuropeptide analysis by MS has seen great advances over the years, the incorporation of more advanced techniques and tools in the future will greatly benefit our understanding of neuropeptides and neurochemical signaling.
Acknowledgements
This review is dedicated to Professor David Lubman for his notable contributions and substantial impact to various fields of mass spectrometry. Preparation of this manuscript was supported in part by the National Science Foundation (CHE- 1710140 and CHE-2108223) and National Institutes of Health through grants (R56 MH110215, R01DK071801 and R01NS029436). A.P. was supported in part by the NIH Chemistry–Biology Interface Training Grant (T32 GM008505). N.V.Q. was supported in part by the National Institutes of Health, under the Ruth L. Kirschstein National Research Service Award from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center (T32 HL007936). A.R.B. would like to thank NIH for a General Medical Sciences National Ruth L. Kirschstein Research Service Award (NRSA) Fellowship 1F31GM119365. L.L. acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy. The authors thank Dr. Erin Gemperline in the Li Research Group for her assistance in graphic design.
Abbreviations:
- DHB
2,5-dihydroxybenzoic acid
- AMT
accurate mass time
- ACN
acetonitrile
- HOAc
acetic acid
- Aβ
amyloid-β protein
- BK
bradykinin
- CE
capillary electrophoresis
- CNS
central nervous system
- CCS
collision cross sections
- CID
collision-induced dissociation
- CCAP
crustacean cardioactive peptide
- cryo-IM-MS
cryogenic IM-MS
- CHCA
α-cyano-4-hydroxy-cinnamic acid
- DAACPs
D-amino acid containing peptides
- DDA
data-dependent acquisition
- DIA
data-independent acquisition
- DESI
desorption electrospray ionization
- DH
diapause hormone
- ExD
electron activated dissociation
- ECD
electron-capture dissociation
- ETD
electron-transfer dissociation
- EThcD
electron-transfer/higher-energy collision dissociation
- ESI
electrospray ionization
- EDTA
ethylenediaminetetraacetic acid
- FDRs
false discovery rates
- FFPE
formaldehyde-fixed paraffin embedding
- FA
formic acid
- FTICR-MS
Fourier-transform ion cyclotron resonance MS
- HCD
high-energy collision dissociation
- HRAM
high-resolution accurate mass
- HCl
hydrochloric acid
- IQ
informed quantitation
- IM-MS
ion mobility MS
- ICAT
isotopic-coded affinity tag
- LFQ
label-free quantitation
- LSI
laserspray ionization
- LC
liquid chromatography
- LESA
liquid extraction surface analysis
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- MALDI
matrix-assisted laser desorption/ionization
- MSH
melanocyte-stimulating hormone
- MeCAT
metal-coded affinity tag
- MeOH
methanol
- MTBE
methyl-tert-butyl ether
- mDa
milliDalton
- M
molarity
- MD
molecular dynamics
- MWCO
molecular weight cut-off
- NIMS
nanostructure initiator MS
- NMR
nuclear magnetic resonance
- OCT
optimal cutting temperature
- PRM
parallel reaction monitoring
- PACAP
pituitary adenylate cyclase activating polypeptide
- pHPMA
poly[N-(2-hydroxypropyl)methacrylamide
- PEI
polyethylenimine
- PTMs
post-translational modifications
- RIAs
radioimmunoassays
- SIMS
secondary ion MS
- SP
single stage MS (MS1) substance P
- MS/MS
tandem MS
- TOF/TOF
tandem TOF
- TOF
time of flight
- TFA
trifluoroacetic acid
Footnotes
Competing Interests
The authors have declared no competing interests.
References
- Adamson KJ, Wang T, Rotgans BA, Kuballa AV, Storey KB, Cummins SF. 2016. Differential peptide expression in the central nervous system of the land snail Theba pisana, between active and aestivated. Peptides 80:61–71. [DOI] [PubMed] [Google Scholar]
- Aerni HR, Cornett DS, Caprioli RM. 2006. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem 78:827–834. [DOI] [PubMed] [Google Scholar]
- Agrawal P, Kumar S, Singh A, Raghava GPS, Singh IK. 2019. NeuroPIpred: a tool to predict, design and scan insect neuropeptides. Scientific Reports 9:5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrends R, Pieper S, Kuhn A, Weisshoff H, Hamester M, Lindemann T, Scheler C, Lehmann K, Taubner K, Linscheid MW. 2007. A metal-coded affinity tag approach to quantitative proteomics. Mol Cell Proteomics 6:1907–1916. [DOI] [PubMed] [Google Scholar]
- Akhtar MN, Southey BR, Andren PE, Sweedler JV, Rodriguez-Zas SL. 2012. Evaluation of database search programs for accurate detection of neuropeptides in tandem mass spectrometry experiments. J Proteome Res 11:6044–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hasani R, Wong JT, Mabrouk OS, McCall JG, Schmitz GP, Porter-Stransky KA, Aragona BJ, Kennedy RT, Bruchas MR. 2018. In vivo detection of optically-evoked opioid peptide release. Elife 7:e36520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alim FZD, Romanova EV, Tay YL, Rahman A, Chan KG, Hong KW, Rogers M, Southey BR, Greenwood MP, Mecawi AS, Mustafa MR, Mahy N, Campbell C, Antunes-Rodrigues J, Sweedler JV, Murphy D, Hindmarch CCT. 2019. Seasonal adaptations of the hypothalamo-neurohypophyseal system of the dromedary camel. PLoS One 14:e0216679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmer J 2011. Algorithms for the de novo sequencing of peptides from tandem mass spectra. Expert Rev Proteomics 8:645–657. [DOI] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. [DOI] [PubMed] [Google Scholar]
- Altelaar AF, Mohammed S, Brans MA, Adan RA, Heck AJ. 2009. Improved identification of endogenous peptides from murine nervous tissue by multiplexed peptide extraction methods and multiplexed mass spectrometric analysis. J Proteome Res 8:870–876. [DOI] [PubMed] [Google Scholar]
- Altelaar AF, van Minnen J, Jimenez CR, Heeren RM, Piersma SR. 2005. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem 77:735–741. [DOI] [PubMed] [Google Scholar]
- Anapindi KDB, Romanova EV, Southey BR, Sweedler JV. 2018. Peptide identifications and false discovery rates using different mass spectrometry platforms. Talanta 182:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Groseclose MR, Deutch AY, Caprioli RM. 2008. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods 5:101–108. [DOI] [PubMed] [Google Scholar]
- Andren PE, Emmett MR, Caprioli RM. 1994. Micro-Electrospray: Zeptomole/attomole per microliter sensitivity for peptides. Journal of the American Society for Mass Spectrometry 5:867–869. [DOI] [PubMed] [Google Scholar]
- Aoki-Kinoshita KF. 2017. A Practical Guide to Using Glycomics Databases. [Google Scholar]
- Atkins N Jr., Ren S, Hatcher N, Burgoon PW, Mitchell JW, Sweedler JV, Gillette MU. 2018. Functional Peptidomics: Stimulus- and Time-of-Day-Specific Peptide Release in the Mammalian Circadian Clock. ACS Chem Neurosci 9:2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerman G, Verleyen P, Clynen E, Huybrechts J, De Loof A, Schoofs L. 2004. Peptidomics. J Chromatogr B Analyt Technol Biomed Life Sci 803:3–16. [DOI] [PubMed] [Google Scholar]
- Bai L, Livnat I, Romanova EV, Alexeeva V, Yau PM, Vilim FS, Weiss KR, Jing J, Sweedler JV. 2013. Characterization of GdFFD, a D-amino acid-containing neuropeptide that functions as an extrinsic modulator of the Aplysia feeding circuit. J Biol Chem 288:32837–32851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Romanova EV, Sweedler JV. 2011. Distinguishing endogenous D-amino acid-containing neuropeptides in individual neurons using tandem mass spectrometry. Anal Chem 83:2794–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluya DL, Garrett TJ, Yost RA. 2007. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal Chem 79:6862–6867. [DOI] [PubMed] [Google Scholar]
- Bardsen K, Gjerstad MD, Partinen M, Kvivik I, Tjensvoll AB, Ruoff P, Omdal R, Brede C. 2019. Considerably Lower Levels of Hypocretin-1 in Cerebrospinal Fluid Is Revealed by a Novel Mass Spectrometry Method Compared with Standard Radioimmunoassay. Anal Chem 91:9323–9329. [DOI] [PubMed] [Google Scholar]
- Bark SJ, Lu WD, Hook V. 2009. Linear and accurate quantitation of proenkephalin-derived peptides by isotopic labeling with internal standards and mass spectrometry. Anal Biochem 389:18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann KY, Church MK, Clough GF, Quist SR, Schmelz M, Skov PS, Anderson CD, Tannert LK, Gimenez-Arnau AM, Frischbutter S, Scheffel J, Maurer M. 2019. Skin microdialysis: methods, applications and future opportunities-an EAACI position paper. Clin Transl Allergy 9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens HL, Chen R, Li L. 2008. Combining microdialysis, NanoLC-MS, and MALDI-TOF/TOF to detect neuropeptides secreted in the crab, Cancer borealis. Anal Chem 80:6949–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens HL, Li L. 2010. Monitoring neuropeptides in vivo via microdialysis and mass spectrometry. Methods Mol Biol 615:57–73. [DOI] [PubMed] [Google Scholar]
- Bemis KD, Harry A, Eberlin LS, Ferreira C, van de Ven SM, Mallick P, Stolowitz M, Vitek O. 2015. Cardinal: an R package for statistical analysis of mass spectrometry-based imaging experiments. Bioinformatics (Oxford, England) 31:2418–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereszczak JZ, Barbu IM, Tan M, Xia M, Jiang X, van Duijn E, Heck AJ. 2012. Structure, stability and dynamics of norovirus P domain derived protein complexes studied by native mass spectrometry. J Struct Biol 177:273–282. [DOI] [PubMed] [Google Scholar]
- Berezovskaya Y, Porrini M, Barran PE. 2013. The effect of salt on the conformations of three model proteins is revealed by variable temperature ion mobility mass spectrometry. Int. J. Mass Spectrom 345:8–18. [Google Scholar]
- Berisha A, Dold S, Guenther S, Desbenoit N, Takats Z, Spengler B, Rompp A. 2014. A comprehensive high-resolution mass spectrometry approach for characterization of metabolites by combination of ambient ionization, chromatography and imaging methods. Rapid Commun Mass Spectrom 28:1779–1791. [DOI] [PubMed] [Google Scholar]
- Berman ESF, Fortson SL, Kulp KS. 2010. Preparation of Single Cells for Imaging Mass Spectrometry. In: Rubakhin SS, Sweedler JV, Editors. Mass Spectrometry Imaging: Principles and Protocols. Totowa: Humana Press Inc. p 253–265. [Google Scholar]
- Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. 2009. Amyloid-beta protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nat Chem 1:326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M, Gupta K, Gowd KH, Balaram P. 2013. Rapid mass spectrometric determination of disulfide connectivity in peptides and proteins. Mol Biosyst 9:1340–1350. [DOI] [PubMed] [Google Scholar]
- Bleiholder C, Dupuis NF, Bowers MT. 2013. Dimerization of chirally mutated Enkephalin neurotransmitters: implications for peptide and protein aggregation mechanisms. J Phys Chem B 117:1770–1779. [DOI] [PubMed] [Google Scholar]
- Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. 2011. Ion mobility–mass spectrometry reveals a conformational conversion from random assembly to β-sheet in amyloid fibril formation. Nat Chem 3:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobba S, Resch GE, Gutheil WG. 2012. A liquid chromatography-tandem mass spectrometry assay for detection and quantitation of the dipeptide Gly-Gln in rat brain. Anal Biochem 425:145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Aye TT, van Veen TA, Heck AJ, Mohammed S. 2008. Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates. Proteomics 8:4624–4632. [DOI] [PubMed] [Google Scholar]
- Boggio KJ, Obasuyi E, Sugino K, Nelson SB, Agar NY, Agar JN. 2011. Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact. Expert Rev Proteomics 8:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongaerts J, Segers K, Van Oudenhove L, Van Wanseele Y, Van Hulle M, De Bundel D, Mangelings D, Smolders I, Vander Heyden Y, Van Eeckhaut A. 2020. A comparative study of UniSpray and electrospray sources for the ionization of neuropeptides in liquid chromatography tandem mass spectrometry. J Chromatogr A 1628:461462. [DOI] [PubMed] [Google Scholar]
- Boonen K, Landuyt B, Baggerman G, Husson SJ, Huybrechts J, Schoofs L. 2008. Peptidomics: the integrated approach of MS, hyphenated techniques and bioinformatics for neuropeptide analysis. J Sep Sci 31:427–445. [DOI] [PubMed] [Google Scholar]
- Bousfield GR, Butnev VY, White WK, Hall AS, Harvey DJ. 2015. Comparison of Follicle-Stimulating Hormone Glycosylation Microheterogenity by Quantitative Negative Mode Nano-Electrospray Mass Spectrometry of Peptide-N Glycanase-Released Oligosaccharides. J Glycomics Lipidomics 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzacco L, Yu H, Zebroski HA, Dengjel J, Deng H, Mojsov S, Steinman RM. 2011. Mass spectrometry analysis and quantitation of peptides presented on the MHC II molecules of mouse spleen dendritic cells. J Proteome Res 10:5016–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand J, Sistla S, Meriaux C, Dorrestein PC, Gaasterland T, Ghassemian M, Wisztorski M, Fournier I, Salzet M, Macagno E, Bafna V. 2011. Automated querying and identification of novel peptides using MALDI mass spectrometric imaging. J Proteome Res 10:1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzzone F, Lectez B, Tollemer H, Leprince J, Dujardin C, Rachidi W, Chatenet D, Baroncini M, Beauvillain JC, Vallarino M, Vaudry H, Chartrel N. 2006. Anatomical distribution and biochemical characterization of the novel RFamide peptide 26RFa in the human hypothalamus and spinal cord. J Neurochem 99:616–627. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Yu Q, Li L. 2015. Advances in Mass Spectrometric Tools for Probing Neuropeptides. Annu. Rev. Anal. Chem 8:485–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger AR, DeLaney K, Johnson J, Li L. 2018. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal Chem 90:240–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger AR, DeLaney K, Liu Y, Vu NQ, Helfenbein K, Li L. 2020a. Mass Spectrometric Profiling of Neuropeptides in Callinectes sapidus during Hypoxia Stress. ACS Chem Neurosci 11:3097–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger AR, Sauer CS, Vu NQ, DeLaney K, Li L. 2020b. Temporal Study of the Perturbation of Crustacean Neuropeptides Due to Severe Hypoxia Using 4-Plex Reductive Dimethylation. J Proteome Res 19:1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger AR, Vu NQ, Johnson J, DeLaney K, Li L. 2020c. A Simple and Effective Sample Preparation Strategy for MALDI-MS Imaging of Neuropeptide Changes in the Crustacean Brain Due to Hypoxia and Hypercapnia Stress. J Am Soc Mass Spectrom 31:1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budamgunta H, Olexiouk V, Luyten W, Schildermans K, Maes E, Boonen K, Menschaert G, Baggerman G. 2018. Comprehensive Peptide Analysis of Mouse Brain Striatum Identifies Novel sORF-Encoded Polypeptides. Proteomics 18:e1700218. [DOI] [PubMed] [Google Scholar]
- Bulbul M, Sinen O, Ozkan A, Aslan MA, Agar A. 2019. Central neuropeptide-S treatment improves neurofunctions of 6-OHDA-induced Parkinsonian rats. Exp Neurol 317:78–86. [DOI] [PubMed] [Google Scholar]
- Burbach JP. 2010. Neuropeptides from concept to online database www.neuropeptides.nl. Eur J Pharmacol 626:27–48. [DOI] [PubMed] [Google Scholar]
- Busby WH Jr., Quackenbush GE, Humm J, Youngblood WW, Kizer JS. 1987. An enzyme(s) that converts glutaminyl-peptides into pyroglutamyl-peptides. Presence in pituitary, brain, adrenal medulla, and lymphocytes. J Biol Chem 262:8532–8536. [PubMed] [Google Scholar]
- Calabrese RL, Norris BJ, Wenning A. 2016. The neural control of heartbeat in invertebrates. Curr Opin Neurobiol 41:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH. 2014. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Research 42:D215–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Gamble AB, Onagi H, Howes J, Hennessy JE, Gu C, Morgan JAM, Easton CJ. 2017. Detection of Biosynthetic Precursors, Discovery of Glycosylated Forms, and Homeostasis of Calcitonin in Human Cancer Cells. Anal Chem 89:6992–6999. [DOI] [PubMed] [Google Scholar]
- Cao Q, Yu Q, Liu Y, Chen Z, Li L. 2020. Signature-Ion-Triggered Mass Spectrometry Approach Enabled Discovery of N- and O-Linked Glycosylated Neuropeptides in the Crustacean Nervous System. J Proteome Res 19:634–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Liu M, Kong S, Wu M, Zhang Y, Yang P. 2021. Recent advances in software tools for more generic and precise intact glycopeptide analysis. Mol Cell Proteomics:100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli RM, Farmer TB, Gile J. 1997. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 69:4751–4760. [DOI] [PubMed] [Google Scholar]
- Casadonte R, Caprioli RM. 2011. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat Protoc 6:1695–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LM, Cavalcanti DM, Araujo CB, Rioli V, Icimoto MY, Gozzo FC, Juliano M, Juliano L, Oliveira V, Ferro ES. 2014. Peptidomic analysis of the neurolysin-knockout mouse brain. J Proteomics 111:238–248. [DOI] [PubMed] [Google Scholar]
- Chansela P, Goto-Inoue N, Zaima N, Sroyraya M, Sobhon P, Setou M. 2012. Visualization of neuropeptides in paraffin-embedded tissue sections of the central nervous system in the decapod crustacean, Penaeus monodon, by imaging mass spectrometry. Peptides 34:10–18. [DOI] [PubMed] [Google Scholar]
- Chapman JD, Goodlett DR, Masselon CD. 2014. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom Rev 33:452–470. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Latham JC, Lane KB, Mobley JA, Polosukhin VV, Wirth PS, Nanney LB, Caprioli RM. 2008. Imaging mass spectrometry of intact proteins from alcohol-preserved tissue specimens: bypassing formalin fixation. J Proteome Res 7:3543–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che FY, Biswas R, Fricker LD. 2005. Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. J Mass Spectrom 40:227–237. [DOI] [PubMed] [Google Scholar]
- Che FY, Fricker LD. 2002. Quantitation of neuropeptides in Cpe(fat)/Cpe(fat) mice using differential isotopic tags and mass spectrometry. Anal Chem 74:3190–3198. [DOI] [PubMed] [Google Scholar]
- Che FY, Lim J, Pan H, Biswas R, Fricker LD. 2005. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics 4:1391–1405. [DOI] [PubMed] [Google Scholar]
- Checco JW, Zhang G, Yuan WD, Le ZW, Jing J, Sweedler JV. 2018. Aplysia allatotropin-related peptide and its newly identified d-amino acid-containing epimer both activate a receptor and a neuronal target. J Biol Chem 293:16862–16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Lietz CB, Li L. 2014. In Situ characterization of proteins using laserspray ionization on a high-performance MALDI-LTQ-Orbitrap mass spectrometer. J Am Soc Mass Spectrom 25:2177–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, OuYang C, Tian Z, Xu M, Li L. 2018. A high resolution atmospheric pressure matrix-assisted laser desorption/ionization-quadrupole-orbitrap MS platform enables in situ analysis of biomolecules by multi-mode ionization and acquisition. Anal Chim Acta 1007:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gao YQ, Russell DH. 2011. How alkali metal ion binding alters the conformation preferences of gramicidin A: a molecular dynamics and ion mobility study. J. Phys. Chem. A 116:689–696. [DOI] [PubMed] [Google Scholar]
- Chen M, Talarovicova A, Zheng Y, Storey KB, Elphick MR. 2019. Neuropeptide precursors and neuropeptides in the sea cucumber Apostichopus japonicus: a genomic, transcriptomic and proteomic analysis. Sci Rep 9:8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Cape SS, Sturm RM, Li L. 2010a. Mass spectrometric imaging of neuropeptides in decapod crustacean neuronal tissues. Methods Mol Biol 656:451–463. [DOI] [PubMed] [Google Scholar]
- Chen R, Hui L, Cape SS, Wang J, Li L. 2010b. Comparative Neuropeptidomic Analysis of Food Intake via a Multi-faceted Mass Spectrometric Approach. ACS Chem Neurosci 1:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hui L, Sturm RM, Li L. 2009a. Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J Am Soc Mass Spectrom 20:1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang X, Conaway MC, Mohtashemi I, Hui L, Viner R, Li L. 2010c. Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster Homarus americanus. J Proteome Res 9:818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Li L. 2010. Mass spectral imaging and profiling of neuropeptides at the organ and cellular domains. Anal Bioanal Chem 397:3185–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Ma M, Hui L, Zhang J, Li L. 2009b. Measurement of neuropeptides in crustacean hemolymph via MALDI mass spectrometry. J Am Soc Mass Spectrom 20:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Xiao M, Buchberger A, Li L. 2014. Quantitative neuropeptidomics study of the effects of temperature change in the crab Cancer borealis. J Proteome Res 13:5767–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RB, Cape SS, Sturm RM, Li L. 2010d. Mass Spectrometric Imaging of Neuropeptides in Decapod Crustacean Neuronal Tissues. In: Rubakhin SS, Sweedler JV, Editors. Mass Spectrometry Imaging: Principles and Protocols. Totowa: Humana Press Inc. p 451–463. [DOI] [PubMed] [Google Scholar]
- Chi H, Sun RX, Yang B, Song CQ, Wang LH, Liu C, Fu Y, Yuan ZF, Wang HP, He SM, Dong MQ. 2010. pNovo: de novo peptide sequencing and identification using HCD spectra. J Proteome Res 9:2713–2724. [DOI] [PubMed] [Google Scholar]
- Chieu HD, Suwansa-Ard S, Wang T, Elizur A, Cummins SF. 2019a. Identification of neuropeptides in the sea cucumber Holothuria leucospilota. Gen Comp Endocrinol 283:113229. [DOI] [PubMed] [Google Scholar]
- Chieu HD, Turner L, Smith MK, Wang T, Nocillado J, Palma P, Suwansa-Ard S, Elizur A, Cummins SF. 2019b. Aquaculture Breeding Enhancement: Maturation and Spawning in Sea Cucumbers Using a Recombinant Relaxin-Like Gonad-Stimulating Peptide. Front Genet 10:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Pennington CL, Wood TD. 2010. Stable isotope labeling method targeting terminal tyrosine for relative peptide quantitation using mass spectrometry. Anal Biochem 401:15–21. [DOI] [PubMed] [Google Scholar]
- Christie AE. 2015. In silico characterization of the neuropeptidome of the Western black widow spider Latrodectus hesperus. Gen Comp Endocrinol 210:63–80. [DOI] [PubMed] [Google Scholar]
- Chung-Davidson YW, Bussy U, Fissette SD, Huerta B, Li W. 2020. Waterborne pheromones modulate gonadotropin-inhibitory hormone levels in sea lamprey (Petromyzon marinus). Gen Comp Endocrinol 288:113358. [DOI] [PubMed] [Google Scholar]
- Cillero-Pastor B, Heeren RM. 2014. Matrix-assisted laser desorption ionization mass spectrometry imaging for peptide and protein analyses: a critical review of on-tissue digestion. J Proteome Res 13:325–335. [DOI] [PubMed] [Google Scholar]
- Clemis EJ, Smith DS, Camenzind AG, Danell RM, Parker CE, Borchers CH. 2012. Quantitation of spatially-localized proteins in tissue samples using MALDI-MRM imaging. Anal Chem 84:3514–3522. [DOI] [PubMed] [Google Scholar]
- Colgrave ML, Xi L, Lehnert SA, Flatscher-Bader T, Wadensten H, Nilsson A, Andren PE, Wijffels G. 2011. Neuropeptide profiling of the bovine hypothalamus: thermal stabilization is an effective tool in inhibiting post-mortem degradation. Proteomics 11:1264–1276. [DOI] [PubMed] [Google Scholar]
- Collier TS, Sarkar P, Franck WL, Rao BM, Dean RA, Muddiman DC. 2010. Direct comparison of stable isotope labeling by amino acids in cell culture and spectral counting for quantitative proteomics. Anal Chem 82:8696–8702. [DOI] [PubMed] [Google Scholar]
- Conant CR, Fuller DR, El-Baba TJ, Zhang Z, Russell DH, Clemmer DE. 2019. Substance P in Solution: Trans-to-Cis Configurational Changes of Penultimate Prolines Initiate Non-enzymatic Peptide Bond Cleavages. J Am Soc Mass Spectrom 30:919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantopoulos TL, Jackson GS, Enke CG. 1999. Effects of salt concentration on analyte response using electrospray ionization mass spectrometry. Journal of the American Society for Mass Spectrometry 10:625–634. [DOI] [PubMed] [Google Scholar]
- Conzelmann M, Williams EA, Krug K, Franz-Wachtel M, Macek B, Jekely G. 2013. The neuropeptide complement of the marine annelid Platynereis dumerilii. BMC Genomics 14:906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley DJ, Hoflack JM, Pelton JT, Saudek V. 1992. Structure of neuropeptide Y dimer in solution. Eur J Biochem 205:1099–1106. [DOI] [PubMed] [Google Scholar]
- Cramer CN, Kelstrup CD, Olsen JV, Haselmann KF, Nielsen PK. 2017. Complete Mapping of Complex Disulfide Patterns with Closely-Spaced Cysteines by In-Source Reduction and Data-Dependent Mass Spectrometry. Anal Chem 89:5949–5957. [DOI] [PubMed] [Google Scholar]
- Crossman L, McHugh NA, Hsieh Y, Korfmacher WA, Chen J. 2006. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun Mass Spectrom 20:284–290. [DOI] [PubMed] [Google Scholar]
- Cui Z, Smith AS. 2019. In vivo measurement of enhanced agouti-related peptide release in the paraventricular nucleus of the hypothalamus through Gs activation of agouti-related peptide neurons. J Biol Methods 6:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, Guerrero A, Parker EA, Robinson RC, Gan J, German JB, Barile D, Lebrilla CB. 2015. Current peptidomics: applications, purification, identification, quantification, and functional analysis. Proteomics 15:1026–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas DC, Martin WF, Hua S, German JB. 2012. Automated glycopeptide analysis--review of current state and future directions. Briefings in Bioinformatics 14:361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haes W, Van Sinay E, Detienne G, Temmerman L, Schoofs L, Boonen K. 2015. Functional neuropeptidomics in invertebrates. Biochim Biophys Acta 1854:812–826. [DOI] [PubMed] [Google Scholar]
- De Sio G, Smith AJ, Galli M, Garancini M, Chinello C, Bono F, Pagni F, Magni F. 2015. A MALDI-Mass Spectrometry Imaging method applicable to different formalin-fixed paraffin-embedded human tissues. Mol Biosyst 11:1507–1514. [DOI] [PubMed] [Google Scholar]
- de Vries MP, Rullens-Ligtvoet N, Hoeberichts WJ, van Gool AJ, van Zeeland MJM, Kouwijzer M, Verheijden GF, Boots AMH, Verhaert PDE, Bos ES. 2005. Identification of Tyrosine-O-Sulfation as post-translational modification of melanoma inhibitory activity (MIA) protein. Biomacromolecular Mass Spectrometry 1:57–73. [Google Scholar]
- DeAtley KL, Colgrave ML, Canovas A, Wijffels G, Ashley RL, Silver GA, Rincon G, Medrano JF, Islas-Trejo A, Fortes MRS, Reverter A, Porto-Neto L, Lehnert SA, Thomas MG. 2018. Neuropeptidome of the Hypothalamus and Pituitary Gland of Indicine x Taurine Heifers: Evidence of Differential Neuropeptide Processing in the Pituitary Gland before and after Puberty. J Proteome Res 17:1852–1865. [DOI] [PubMed] [Google Scholar]
- Delafield DG, Li L. 2020. Recent Advances in Analytical Approaches for Glycan and Glycopeptide Quantitation. Mol Cell Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaney K, Buchberger A, Li L. 2018. Identification, Quantitation, and Imaging of the Crustacean Peptidome. Methods Mol Biol 1719:247–269. [DOI] [PubMed] [Google Scholar]
- DeLaney K, Buchberger AR, Atkinson L, Grunder S, Mousley A, Li L. 2018. New techniques, applications and perspectives in neuropeptide research. J Exp Biol 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaney K, Cao W, Ma Y, Ma M, Zhang Y, Li L. 2020. PRESnovo: Prescreening Prior to de novo Sequencing to Improve Accuracy and Sensitivity of Neuropeptide Identification. J Am Soc Mass Spectrom 31:1358–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaney K, Li L. 2019a. Capillary electrophoresis coupled to MALDI mass spectrometry imaging with large volume sample stacking injection for improved coverage of C. borealis neuropeptidome. Analyst 145:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaney K, Li L. 2019b. Data Independent Acquisition Mass Spectrometry Method for Improved Neuropeptidomic Coverage in Crustacean Neural Tissue Extracts. Anal Chem 91:5150–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaney K, Li L. 2020. Neuropeptidomic Profiling and Localization in the Crustacean Cardiac Ganglion Using Mass Spectrometry Imaging with Multiple Platforms. J. Am. Chem. Soc 31:2469–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaney K, Phetsanthad A, Li L. 2020. Advances in High-Resolution Maldi Mass Spectrometry for Neurobiology. Mass Spectrometry Reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliconstantinos G, Barton S, Soloviev M, Page N. 2017. Mouse Hemokinin-1 Decapeptide Subjected to a Brain-specific Post-translational Modification. In Vivo 31:991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio DM, Kusmierz JJ, Zhu X, Dass C, Hilton D, Robertson JT, Sacks HS. 1993. Mass spectrometric analysis of opioid and tachykinin neuropeptides in non-secreting and ACTH-secreting human pituitary adenomas. Biol Mass Spectrom 22:89–97. [DOI] [PubMed] [Google Scholar]
- Desiderio DM, Yamada S. 1982. Measurement of endogenous leucine enkephalin in canine thalamus by high-performance liquid chromatography and field desorption mass spectrometry. Journal of Chromatography A 239:87–95. [DOI] [PubMed] [Google Scholar]
- Dewez F, De Pauw E, Heeren RMA, Balluff B. 2021. Multilabel Per-Pixel Quantitation in Mass Spectrometry Imaging. Anal Chem 93:1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Y, Zhang Y, Zhang L, Tao T, Lu H. 2017. MdFDIA: A Mass Defect Based Four-Plex Data-Independent Acquisition Strategy for Proteome Quantification. Anal Chem 89:10248–10255. [DOI] [PubMed] [Google Scholar]
- Dickinson PS, Qu X, Stanhope ME. 2016. Neuropeptide modulation of pattern-generating systems in crustaceans: comparative studies and approaches. Curr Opin Neurobiol 41:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PS, Sreekrishnan A, Kwiatkowski MA, Christie AE. 2015. Distinct or shared actions of peptide family isoforms: I. Peptide-specific actions of pyrokinins in the lobster cardiac neuromuscular system. J Exp Biol 218:2892–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl HC, Beine B, Elm J, Trede D, Ahrens M, Eisenacher M, Marcus K, Meyer HE, Henkel C. 2015. The challenge of on-tissue digestion for MALDI MSI- a comparison of different protocols to improve imaging experiments. Anal Bioanal Chem 407:2223–2243. [DOI] [PubMed] [Google Scholar]
- Diesner M, Predel R, Neupert S. 2018. Neuropeptide Mapping of Dimmed Cells of Adult Drosophila Brain. J Am Soc Mass Spectrom 29:890–902. [DOI] [PubMed] [Google Scholar]
- Do TD, Checco JW, Tro M, Shea JE, Bowers MT, Sweedler JV. 2018a. Conformational investigation of the structure-activity relationship of GdFFD and its analogues on an achatin-like neuropeptide receptor of Aplysia californica involved in the feeding circuit. Phys Chem Chem Phys 20:22047–22057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do TD, Ellis JF, Neumann EK, Comi TJ, Tillmaand EG, Lenhart AE, Rubakhin SS, Sweedler JV. 2018b. Optically Guided Single Cell Mass Spectrometry of Rat Dorsal Root Ganglia to Profile Lipids, Peptides and Proteins. Chemphyschem 19:1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do TD, LaPointe NE, Sangwan S, Teplow DB, Feinstein SC, Sawaya MR, Eisenberg DS, Bowers MT. 2014. Factors that drive peptide assembly from native to amyloid structures: experimental and theoretical analysis of [Leu-5]-enkephalin mutants. J. Phys. Chem. B 118:7247–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zhang Y, Meng Z, Zhu X, Gan H, Gu R, Wu Z, Li J, Zheng Y, Yang B, Dou G. 2018. A LC-MS/MS method to monitor the concentration of HYD-PEP06, a RGD-modified Endostar mimetic peptide in rat blood. J Chromatogr B Analyt Technol Biomed Life Sci 1092:296–305. [DOI] [PubMed] [Google Scholar]
- Dowell JA, Heyden WV, Li L. 2006. Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: Enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J Proteome Res 5:3368–3375. [DOI] [PubMed] [Google Scholar]
- Dreisewerd K 2014. Recent methodological advances in MALDI mass spectrometry. Anal Bioanal Chem 406:2261–2278. [DOI] [PubMed] [Google Scholar]
- Dueñas ME, Essner JJ, Lee YJ. 2017. 3D MALDI Mass Spectrometry Imaging of a Single Cell: Spatial Mapping of Lipids in the Embryonic Development of Zebrafish. Scientific Reports 7:14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergin B, Meding S, Langer R, Kap M, Viertler C, Schott C, Ferch U, Riegman P, Zatloukal K, Walch A, Becker KF. 2010. Proteomic analysis of PAXgene-fixed tissues. J Proteome Res 9:5188–5196. [DOI] [PubMed] [Google Scholar]
- Eriksson C, Masaki N, Yao I, Hayasaka T, Setou M. 2013. MALDI Imaging Mass Spectrometry-A Mini Review of Methods and Recent Developments. Mass Spectrom (Tokyo) 2:S0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falth M, Skold K, Norrman M, Svensson M, Fenyo D, Andren PE. 2006. SwePep, a database designed for endogenous peptides and mass spectrometry. Mol Cell Proteomics 5:998–1005. [DOI] [PubMed] [Google Scholar]
- Fernandes SV. 2004. Nasal fractures. ANZ J Surg 74:285; author reply 285–286. [DOI] [PubMed] [Google Scholar]
- Fleites LA, Johnson R, Kruse AR, Nachman RJ, Hall DG, MacCoss M, Heck ML. 2020. Peptidomics Approaches for the Identification of Bioactive Molecules from Diaphorina citri. J Proteome Res 19:1392–1408. [DOI] [PubMed] [Google Scholar]
- Flintegaard TV, Thygesen P, Rahbek-Nielsen H, Levery SB, Kristensen C, Clausen H, Bolt G. 2010. N-glycosylation increases the circulatory half-life of human growth hormone. Endocrinology 151:5326–5336. [DOI] [PubMed] [Google Scholar]
- Fort KL, Silveira JA, Pierson NA, Servage KA, Clemmer DE, Russell DH. 2014. From Solution to the Gas Phase: Factors That Influence Kinetic Trapping of Substance P in the Gas Phase. J. Phys. Chem. B 118:14336–14344. [DOI] [PubMed] [Google Scholar]
- Franck J, Arafah K, Barnes A, Wisztorski M, Salzet M, Fournier I. 2009. Improving tissue preparation for matrix-assisted laser desorption ionization mass spectrometry imaging. Part 1: using microspotting. Anal Chem 81:8193–8202. [DOI] [PubMed] [Google Scholar]
- Fredrick WS, Ravichandran S. 2012. Hemolymph proteins in marine crustaceans. Asian Pacific Journal of Tropical Biomedicine 2:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey BL, Ladror DT, Sondalle SB, Krusemark CJ, Jue AL, Coon JJ, Smith LM. 2013. Chemical derivatization of peptide carboxyl groups for highly efficient electron transfer dissociation. J Am Soc Mass Spectrom 24:1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker L 2018. Quantitative Peptidomics: General Considerations. Methods Mol Biol 1719:121–140. [DOI] [PubMed] [Google Scholar]
- Fricker LD. 2006. Neuropeptidomics of drug abuse: Quantitation and identification of neuropeptides using mass spectrometry. Neuropeptides 40:149–149. [Google Scholar]
- Fricker LD, Lim J, Pan H, Che FY. 2006. Peptidomics: identification and quantification of endogenous peptides in neuroendocrine tissues. Mass Spectrom Rev 25:327–344. [DOI] [PubMed] [Google Scholar]
- Fridjonsdottir E, Nilsson A, Wadensten H, Andrén PE. 2018. Brain Tissue Sample Stabilization and Extraction Strategies for Neuropeptidomics. In: Schrader M, Fricker L, Editors. Peptidomics: Methods and Strategies. New York, NY: Springer New York. p 41–49. [DOI] [PubMed] [Google Scholar]
- Friesen WL, Schultz BJ, Destino JF, Alivio TE, Steet JR, Banerjee S, Wood TD. 2015. Two-dimensional graphene as a matrix for MALDI imaging mass spectrometry. J Am Soc Mass Spectrom 26:1963–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost DC, Buchberger AR, Li L. 2017. Mass Defect-Based Dimethyl Pyrimidinyl Ornithine (DiPyrO) Tags for Multiplex Quantitative Proteomics. Analytical Chemistry 89:10798–10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost DC, Feng Y, Li L. 2020. 21-plex DiLeu Isobaric Tags for High-Throughput Quantitative Proteomics. Anal Chem 92:8228–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost DC, Greer T, Li L. 2015. High-resolution enabled 12-plex DiLeu isobaric tags for quantitative proteomics. Anal Chem 87:1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost DC, Greer T, Xiang F, Liang Z, Li L. 2015. Development and characterization of novel 8-plex DiLeu isobaric labels for quantitative proteomics and peptidomics. Rapid Commun Mass Spectrom 29:1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li L. 2005. De novo sequencing of neuropeptides using reductive isotopic methylation and investigation of ESI QTOF MS/MS fragmentation pattern of neuropeptides with N-terminal dimethylation. Anal Chem 77:7783–7795. [DOI] [PubMed] [Google Scholar]
- Gade G, Marco HG. 2015. The decapod red pigment-concentrating hormone (Panbo-RPCH) is the first identified neuropeptide of the order Plecoptera and is interpreted as homoplastic character state. Gen Comp Endocrinol 221:228–235. [DOI] [PubMed] [Google Scholar]
- Gatenholm B, Gobom J, Skillback T, Blennow K, Zetterberg H, Brittberg M. 2019. Peptidomic analysis of cartilage and subchondral bone in OA patients. Eur J Clin Invest 49:e13082. [DOI] [PubMed] [Google Scholar]
- Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. 2010. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods 7:383–385. [DOI] [PubMed] [Google Scholar]
- Gemperline E, Chen B, Li L. 2014. Challenges and recent advances in mass spectrometric imaging of neurotransmitters. Bioanalysis 6:525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperline E, Rawson S, Li L. 2014. Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal Chem 86:10030–10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidden J, Bowers M. 2002. Gas-phase conformational and energetic properties of deprotonated dinucleotides. The European Physical Journal D-Atomic, Molecular, Optical and Plasma Physics 20:409–419. [Google Scholar]
- Gidden J, Bushnell JE, Bowers MT. 2001. Gas-phase conformations and folding energetics of oligonucleotides: dTG-and dGT. J. Am. Chem. Soc 123:5610–5611. [DOI] [PubMed] [Google Scholar]
- Glover MS, Bellinger EP, Radivojac P, Clemmer DE. 2015. Penultimate proline in neuropeptides. Anal Chem 87:8466–8472. [DOI] [PubMed] [Google Scholar]
- Goettig P 2016. Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases. Int J Mol Sci 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ramos MDM, Gomez Ramos MJ, Martinez Galera M, Gil Garcia MD, Fernandez-Alba AR. 2018. Analysis and evaluation of (neuro)peptides in honey bees exposed to pesticides in field conditions. Environ Pollut 235:750–760. [DOI] [PubMed] [Google Scholar]
- Goodwin RJ, Dungworth JC, Cobb SR, Pitt AR. 2008. Time-dependent evolution of tissue markers by MALDI-MS imaging. Proteomics 8:3801–3808. [DOI] [PubMed] [Google Scholar]
- Goodwin RJ, Lang AM, Allingham H, Boren M, Pitt AR. 2010. Stopping the clock on proteomic degradation by heat treatment at the point of tissue excision. Proteomics 10:1751–1761. [DOI] [PubMed] [Google Scholar]
- Goodwin RJA. 2012. Sample preparation for mass spectrometry imaging: small mistakes can lead to big consequences. J Proteomics 75:4893–4911. [DOI] [PubMed] [Google Scholar]
- Goodwin RJA, Nilsson A, Borg D, Langridge-Smith PRR, Harrison DJ, Mackay CL, Iverson SL, Andren PE. 2012. Conductive carbon tape used for support and mounting of both whole animal and fragile heat-treated tissue sections for MALDI MS imaging and quantitation. J Proteomics 75:4912–4920. [DOI] [PubMed] [Google Scholar]
- Greer T, Lietz CB, Xiang F, Li L. 2015. Novel isotopic N,N-dimethyl leucine (iDiLeu) reagents enable absolute quantification of peptides and proteins using a standard curve approach. J Am Soc Mass Spectrom 26:107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. 2007. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom 42:254–262. [DOI] [PubMed] [Google Scholar]
- Groseclose MR, Castellino S. 2013. A mimetic tissue model for the quantification of drug distributions by MALDI imaging mass spectrometry. Anal Chem 85:10099–10106. [DOI] [PubMed] [Google Scholar]
- Grund T, Goyon S, Li Y, Eliava M, Liu H, Charlet A, Grinevich V, Neumann ID. 2017. Neuropeptide S Activates Paraventricular Oxytocin Neurons to Induce Anxiolysis. J Neurosci 37:12214–12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther S, Römpp A, Kummer W, Spengler B. 2011. AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5μm spatial resolution and high mass accuracy. Int. J. Mass Spectrom 305:228–237. [Google Scholar]
- Guvenc-Bayram G, Altinbas B, Iqbal A, Cerci E, Udum D, Yilmaz MS, Erdost H, Yalcin-Ulger E, Ilhan T, Ersoy F, Uz E, Yalcin M. 2020. Intracerebroventricularly injected nesfatin-1 activates central cyclooxygenase and lipoxygenase pathways. Auton Neurosci 226:102670. [DOI] [PubMed] [Google Scholar]
- Guzman-Ruiz MA, Ramirez-Corona A, Guerrero-Vargas NN, Sabath E, Ramirez-Plascencia OD, Fuentes-Romero R, Leon-Mercado LA, Basualdo Sigales M, Escobar C, Buijs RM. 2015. Role of the Suprachiasmatic and Arcuate Nuclei in Diurnal Temperature Regulation in the Rat. J Neurosci 35:15419–15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale OJ, Ryumin P, Brown JM, Morris M, Cramer R. 2021. Production and analysis of multiply charged negative ions by liquid atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry. Rapid Communications in Mass Spectrometry 35:e8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim A, Westerlind U, Pett C, Schorlemer M, Ruetschi U, Brinkmalm G, Sihlbom C, Lengqvist J, Larson G, Nilsson J. 2014. Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J Proteome Res 13:6024–6032. [DOI] [PubMed] [Google Scholar]
- Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos JM, Bouzom F, Stauber J. 2012. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J Proteomics 75:4952–4961. [DOI] [PubMed] [Google Scholar]
- Han B, Fang Y, Feng M, Hu H, Qi Y, Huo X, Meng L, Wu B, Li J. 2015. Quantitative Neuropeptidome Analysis Reveals Neuropeptides Are Correlated with Social Behavior Regulation of the Honeybee Workers. J Proteome Res 14:4382–4393. [DOI] [PubMed] [Google Scholar]
- Hanrieder J, Ljungdahl A, Andersson M. 2012. MALDI imaging mass spectrometry of neuropeptides in Parkinson's disease. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrieder J, Malmberg P, Ewing AG. 2015. Spatial neuroproteomics using imaging mass spectrometry. Biochim Biophys Acta 1854:718–731. [DOI] [PubMed] [Google Scholar]
- Hansen LH, Madsen TD, Goth CK, Clausen H, Chen Y, Dzhoyashvili N, Iyer SR, Sangaralingham SJ, Burnett JC Jr., Rehfeld JF, Vakhrushev SY, Schjoldager KT, Goetze JP. 2019. Discovery of O-glycans on atrial natriuretic peptide (ANP) that affect both its proteolytic degradation and potency at its cognate receptor. J Biol Chem 294:12567–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K, Tu X, Ullah H, McNeill MR, Zhang Z. 2019. Novel Lom-dh Genes Play Potential Role in Promoting Egg Diapause of Locusta migratoria L. Front Physiol 10:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Johnson J, Lietz CB, Buchberger A, Frost D, Kao WJ, Li L. 2017. Mass Defect-Based N,N-Dimethyl Leucine Labels for Quantitative Proteomics and Amine Metabolomics of Pancreatic Cancer Cells. Analytical Chemistry 89:1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MM, Alam MA, Shoombuatong W, Deng H-W, Manavalan B, Kurata H. 2021. NeuroPred-FRL: an interpretable prediction model for identifying neuropeptide using feature representation learning. Briefings in Bioinformatics. [DOI] [PubMed] [Google Scholar]
- Hayakawa E, Watanabe H, Menschaert G, Holstein TW, Baggerman G, Schoofs L. 2019. A combined strategy of neuropeptide prediction and tandem mass spectrometry identifies evolutionarily conserved ancient neuropeptides in the sea anemone Nematostella vectensis. PLoS One 14:e0215185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Merrill AE, Bailey DJ, Still AJ, Westphall MS, Strieter ER, Pagliarini DJ, Coon JJ. 2013a. Neutron-encoded mass signatures for multiplexed proteome quantification. Nat Methods 10:332–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Merrill AE, Stefely JA, Bailey DJ, Wenger CD, Westphall MS, Pagliarini DJ, Coon JJ. 2013b. Amine-reactive neutron-encoded labels for highly plexed proteomic quantitation. Mol Cell Proteomics 12:3360–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. 2014. The one hour yeast proteome. Mol Cell Proteomics 13:339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck AJ. 2008. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods 5:927–933. [DOI] [PubMed] [Google Scholar]
- Heijs B, Tolner EA, Bovee JV, van den Maagdenberg AM, McDonnell LA. 2015. Brain Region-Specific Dynamics of On-Tissue Protein Digestion Using MALDI Mass Spectrometry Imaging. J Proteome Res 14:5348–5354. [DOI] [PubMed] [Google Scholar]
- Herbert Z, Rauser S, Williams L, Kapan N, Guntner M, Walch A, Boyan G. 2010. Developmental expression of neuromodulators in the central complex of the grasshopper Schistocerca gregaria. J Morphol 271:1509–1526. [DOI] [PubMed] [Google Scholar]
- Herlenius E, Lagercrantz H. 2004. Development of neurotransmitter systems during critical periods. Exp Neurol 190 Suppl 1:S8–21. [DOI] [PubMed] [Google Scholar]
- Hokfelt T, Broberger C, Xu ZQ, Sergeyev V, Ubink R, Diez M. 2000. Neuropeptides--an overview. Neuropharmacology 39:1337–1356. [DOI] [PubMed] [Google Scholar]
- Hook V, Lietz CB, Podvin S, Cajka T, Fiehn O. 2018. Diversity of Neuropeptide Cell-Cell Signaling Molecules Generated by Proteolytic Processing Revealed by Neuropeptidomics Mass Spectrometry. J Am Soc Mass Spectrom 29:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Xie F, Sweedler JV. 2012. Relative quantitation of neuropeptides over a thousand-fold concentration range. J Am Soc Mass Spectrom 23:2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CC, Baker MW, Gaasterland T, Meehan MJ, Macagno ER, Dorrestein PC. 2017. Top-Down Atmospheric Ionization Mass Spectrometry Microscopy Combined With Proteogenomics. Anal Chem 89:8251–8258. [DOI] [PubMed] [Google Scholar]
- Hsu JL, Huang SY, Chen SH. 2006. Dimethyl multiplexed labeling combined with microcolumn separation and MS analysis for time course study in proteomics. Electrophoresis 27:3652–3660. [DOI] [PubMed] [Google Scholar]
- Hsu JL, Huang SY, Shiea JT, Huang WY, Chen SH. 2005. Beyond quantitative proteomics: signal enhancement of the a1 ion as a mass tag for peptide sequencing using dimethyl labeling. J Proteome Res 4:101–108. [DOI] [PubMed] [Google Scholar]
- Hui L, Cunningham R, Zhang Z, Cao W, Jia C, Li L. 2011. Discovery and characterization of the Crustacean hyperglycemic hormone precursor related peptides (CPRP) and orcokinin neuropeptides in the sinus glands of the blue crab Callinectes sapidus using multiple tandem mass spectrometry techniques. J Proteome Res 10:4219–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, D'Andrea BT, Jia C, Liang Z, Christie AE, Li L. 2013. Mass spectrometric characterization of the neuropeptidome of the ghost crab Ocypode ceratophthalma (Brachyura, Ocypodidae). Gen Comp Endocrinol 184:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, Xiang F, Zhang Y, Li L. 2012. Mass spectrometric elucidation of the neuropeptidome of a crustacean neuroendocrine organ. Peptides 36:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme H, Fridjonsdottir E, Gunnarsdottir H, Vallianatou T, Zhang X, Wadensten H, Shariatgorji R, Nilsson A, Bezard E, Svenningsson P, Andren PE. 2020. Simultaneous mass spectrometry imaging of multiple neuropeptides in the brain and alterations induced by experimental parkinsonism and L-DOPA therapy. Neurobiol Dis 137:104738. [DOI] [PubMed] [Google Scholar]
- Hummon AB, Hummon NP, Corbin RW, Li L, Vilim FS, Weiss KR, Sweedler JV. 2003. From precursor to final peptides: a statistical sequence-based approach to predicting prohormone processing. J Proteome Res 2:650–656. [DOI] [PubMed] [Google Scholar]
- Insel TR. 2010. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron 65:768–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inutan ED, Richards AL, Wager-Miller J, Mackie K, McEwen CN, Trimpin S. 2011. Laserspray ionization, a new method for protein analysis directly from tissue at atmospheric pressure with ultrahigh mass resolution and electron transfer dissociation. Mol Cell Proteomics 10:M110 000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inutan ED, Wager-Miller J, Mackie K, Trimpin S. 2012. Laserspray ionization imaging of multiply charged ions using a commercial vacuum MALDI ion source. Anal Chem 84:9079–9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inutan ED, Wang B, Trimpin S. 2011. Commercial intermediate pressure MALDI ion mobility spectrometry mass spectrometer capable of producing highly charged laserspray ionization ions. Anal Chem 83:678–684. [DOI] [PubMed] [Google Scholar]
- Jarecki JL, Frey BL, Smith LM, Stretton AO. 2011. Discovery of neuropeptides in the nematode Ascaris suum by database mining and tandem mass spectrometry. J Proteome Res 10:3098–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarecki JL, Viola IR, Andersen KM, Miller AH, Ramaker MA, Vestling MM, Stretton AO. 2013. Three independent techniques localize expression of transcript afp-11 and its bioactive peptide products to the paired AVK neurons in Ascaris suum: in situ hybridization, immunocytochemistry, and single cell mass spectrometry. ACS Chem Neurosci 4:418–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrychowski MP, Huttlin EL, Haas W, Sowa ME, Rad R, Gygi SP. 2011. Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol Cell Proteomics 10:M111 009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner M, Ellis J, Huang WC, Lloyd Raven E, Roberts GC, Oldham NJ. 2011. Detection of a protein conformational equilibrium by electrospray ionisation-ion mobility-mass spectrometry. Angew Chem Int Ed Engl 50:8291–8294. [DOI] [PubMed] [Google Scholar]
- Jeong K, Kim S, Bandeira N. 2012. False discovery rates in spectral identification. BMC Bioinformatics 13 Suppl 16:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K, Kim S, Pevzner PA. 2013. UniNovo: a universal tool for de novo peptide sequencing. Bioinformatics 29:1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Hui L, Cao W, Lietz CB, Jiang X, Chen R, Catherman AD, Thomas PM, Ge Y, Kelleher NL, Li L. 2012. High-definition de novo sequencing of crustacean hyperglycemic hormone (CHH)-family neuropeptides. Mol Cell Proteomics 11:1951–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Lietz CB, Yu Q, Li L. 2014. Site-specific characterization of (D)-amino acid containing peptide epimers by ion mobility spectrometry. Anal Chem 86:2972–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Lietz CB, Yu Q, Li L. 2016. Site-specific Localization of D-Amino Acids in Bioactive Peptides by Ion Mobility SpectrometryAnalysis of Post-Translational Modifications and Proteolysis in Neuroscience. p 43–53. [Google Scholar]
- Jiang H, Favaro E, Goulbourne CN, Rakowska PD, Hughes GM, Ryadnov MG, Fong LG, Young SG, Ferguson DJ, Harris AL, Grovenor CR. 2014. Stable isotope imaging of biological samples with high resolution secondary ion mass spectrometry and complementary techniques. Methods 68:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Liang Z, Hao L, Li L. 2016. Investigation of signaling molecules and metabolites found in crustacean hemolymph via in vivo microdialysis using a multifaceted mass spectrometric platform. Electrophoresis 37:1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Chen R, Wang J, Metzler A, Tlusty M, Li L. 2012. Mass spectral charting of neuropeptidomic expression in the stomatogastric ganglion at multiple developmental stages of the lobster Homarus americanus. ACS Chem Neurosci 3:439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Miao A, Zhang X, Cai Y, Lu Y, Zhang Y, Lu H. 2013. Realization of on-tissue protein identification by highly efficient in situ digestion with graphene-immobilized trypsin for MALDI imaging analysis. Analyst 138:1645–1648. [DOI] [PubMed] [Google Scholar]
- Jo K, Heien ML, Thompson LB, Zhong M, Nuzzo RG, Sweedler JV. 2007. Mass spectrometric imaging of peptide release from neuronal cells within microfluidic devices. Lab Chip 7:1454–1460. [DOI] [PubMed] [Google Scholar]
- Johansson C, Samskog J, Sundstrom L, Wadensten H, Bjorkesten L, Flensburg J. 2006. Differential expression analysis of Escherichia coli proteins using a novel software for relative quantitation of LC-MS/MS data. Proteomics 6:4475–4485. [DOI] [PubMed] [Google Scholar]
- Jones EA, Shyti R, van Zeijl RJM, van Heiningen SH, Ferrari MD, Deelder AM, Tolner EA, van den Maagdenberg A, McDonnell LA. 2012. Imaging mass spectrometry to visualize biomolecule distributions in mouse brain tissue following hemispheric cortical spreading depression. J Proteomics 75:5027–5035. [DOI] [PubMed] [Google Scholar]
- Jurneczko E, Barran PE. 2011. How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst 136:20–28. [DOI] [PubMed] [Google Scholar]
- Kaddis CS, Loo JA. 2007. Native protein MS and ion mobility large flying proteins with ESI. Anal Chem 79:1778–1784. [DOI] [PubMed] [Google Scholar]
- Kaletaş BK, van der Wiel IM, Stauber J, Dekker LJ, Güzel C, Kros JM, Luider TM, Heeren RMA. 2009. Sample preparation issues for tissue imaging by imaging MS. Proteomics 9:2622–2633. [DOI] [PubMed] [Google Scholar]
- Källback P, Nilsson A, Shariatgorji M, Andrén PE. 2016. msIQuant – Quantitation Software for Mass Spectrometry Imaging Enabling Fast Access, Visualization, and Analysis of Large Data Sets. Analytical Chemistry 88:4346–4353. [DOI] [PubMed] [Google Scholar]
- Kallback P, Shariatgorji M, Nilsson A, Andren PE. 2012. Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J Proteomics 75:4941–4951. [DOI] [PubMed] [Google Scholar]
- Kang J, Fang Y, Yao P, Li N, Tang Q, Huang J. 2019. NeuroPP: A Tool for the Prediction of Neuropeptide Precursors Based on Optimal Sequence Composition. Interdiscip Sci 11:108–114. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Soderstrom M, Fenyo D, Nilsson A, Falth M, Skold K, Svensson M, Pettersen H, Lindqvist S, Svenningsson P, Andren PE, Bjorkesten L. 2007. An automated method for scanning LC-MS data sets for significant peptides and proteins, including quantitative profiling and interactive confirmation. J Proteome Res 6:2888–2895. [DOI] [PubMed] [Google Scholar]
- Katali OKH, Marco HG, Gade G. 2020. Structure-Activity Studies on the Hypertrehalosemic Hormone II of the Stick Insect Carausius morosus (Phasmatodea): Carbohydrate-Mobilization and Cardio-Stimulatory Activities. Front Physiol 11:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Wei P, Wancewicz B, Cross TL, Rey FE, Li L. 2020. Extraction optimization for combined metabolomics, peptidomics, and proteomics analysis of gut microbiota samples. J Mass Spectrom n/a:e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz V, Calligaris D, Feldman DR, Changelian A, Laws ER, Santagata S, Agar NY, Van Berkel GJ. 2015. Profiling of adrenocorticotropic hormone and arginine vasopressin in human pituitary gland and tumor thin tissue sections using droplet-based liquid-microjunction surface-sampling-HPLC-ESI-MS-MS. Anal Bioanal Chem 407:5989–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Kim EJ, Go HJ, Oh HY, Lin M, Elphick MR, Park NG. 2016. Identification of a novel starfish neuropeptide that acts as a muscle relaxant. J Neurochem 137:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Shin M, Song JS, An S, Kim HJ. 2011a. C-terminal de novo sequencing of peptides using oxazolone-based derivatization with bromine signature. Anal Biochem 419:211–216. [DOI] [PubMed] [Google Scholar]
- Kim Y, Bark S, Hook V, Bandeira N. 2011b. NeuroPedia: neuropeptide database and spectral library. Bioinformatics 27:2772–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan P, Kay RG, Brouwers B, Herranz-Perez V, Jura M, Larraufie P, Jerber J, Pembroke J, Bartels T, White A, Gribble FM, Reimann F, Farooqi IS, O'Rahilly S, Merkle FT. 2018. Quantitative mass spectrometry for human melanocortin peptides in vitro and in vivo suggests prominent roles for beta-MSH and desacetyl alpha-MSH in energy homeostasis. Mol Metab 17:82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Carvalho L, Zaia J. 2018. Application of network smoothing to glycan LC-MS profiling. Bioinformatics 34:3511–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Zaia J. 2019. glypy: An Open Source Glycoinformatics Library. J Proteome Res 18:3532–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeniger SL, Talaty N, Luo Y, Ready D, Voorbach M, Seifert T, Cepa S, Fagerland JA, Bouska J, Buck W, Johnson RW, Spanton S. 2011. A quantitation method for mass spectrometry imaging. Rapid Commun Mass Spectrom 25:503–510. [DOI] [PubMed] [Google Scholar]
- Kompauer M, Heiles S, Spengler B. 2017. Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-μm lateral resolution. Nat Methods 14:90–96. [DOI] [PubMed] [Google Scholar]
- Kondrat FD, Kowald GR, Scarff CA, Scrivens JH, Blindauer CA. 2013. Resolution of a paradox by native mass spectrometry: facile occupation of all four metal binding sites in the dimeric zinc sensor SmtB. Chem Commun 49:813–815. [DOI] [PubMed] [Google Scholar]
- Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. 2017. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods 14:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig S, Bayer M, Marco H, Gade G. 2019. The hypertrehalosaemic neuropeptide conformational twins of cicadas consist of only L-amino acids: are they cis-trans isomers? Amino Acids 51:1023–1028. [DOI] [PubMed] [Google Scholar]
- Konig S, Marco H, Gade G. 2017. The hypertrehalosemic neuropeptides of cicadas are structural isomers-evidence by ion mobility mass spectrometry. Anal Bioanal Chem 409:6415–6420. [DOI] [PubMed] [Google Scholar]
- Konijnenberg A, Butterer A, Sobott F. 2013. Native ion mobility-mass spectrometry and related methods in structural biology. Biochim Biophys Acta 1834:1239–1256. [DOI] [PubMed] [Google Scholar]
- Kovacs AK, Atlasz T, Werling D, Szabo E, Reglodi D, Toth GK. 2020. Stability Test of PACAP in Eye Drops. J Mol Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. 2008. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134:353–364. [DOI] [PubMed] [Google Scholar]
- Kunz TO, Chen J, Megha, Wegener C. 2018. Metabolic Labeling to Quantify Drosophila Neuropeptides and Peptide Hormones. Methods Mol Biol 1719:175–185. [DOI] [PubMed] [Google Scholar]
- Kurian JR, Keen KL, Kenealy BP, Garcia JP, Hedman CJ, Terasawa E. 2015. Acute Influences of Bisphenol A Exposure on Hypothalamic Release of Gonadotropin-Releasing Hormone and Kisspeptin in Female Rhesus Monkeys. Endocrinology 156:2563–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushikata T, Hirota K. 2011. Neuropeptide microdialysis in free-moving animals. Methods Mol Biol 789:261–269. [DOI] [PubMed] [Google Scholar]
- Kusmierz JJ, Desiderio DM. 1992. Characterization of an aminopeptidase in cerebrospinal fluid. Structure elucidation of enzyme hydrolysis products of synthetic methionine-enkephalin by reversed-phase high-performance liquid chromatography and mass spectrometry. J Chromatogr 574:189–196. [PubMed] [Google Scholar]
- Kwok KY, Choi TLS, Kwok WH, Lau MY, Leung EMK, Leung GNW, Wong JKY, Wan TSM, Adrian FF, Prabhu A, Ho ENM. 2020. Detection of bioactive peptides including gonadotrophin-releasing factors (GnRHs) in horse urine using ultra-high performance liquid chromatography-high resolution mass spectrometry (UHPLC/HRMS). Drug Test Anal. [DOI] [PubMed] [Google Scholar]
- Lamont L, Baumert M, Ogrinc Potocnik N, Allen M, Vreeken R, Heeren RMA, Porta T. 2017. Integration of Ion Mobility MS(E) after Fully Automated, Online, High-Resolution Liquid Extraction Surface Analysis Micro-Liquid Chromatography. Anal Chem 89:11143–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa MS, Garcia SI, Schuman ML, Peres Diaz LS, Aisicovich M, Pirola CJ. 2020. Cardiovascular and body weight regulation changes in transgenic mice overexpressing thyrotropin-releasing hormone (TRH). J Physiol Biochem 76:599–608. [DOI] [PubMed] [Google Scholar]
- Lanekoff I, Laskin J. 2017. Quantitative mass spectrometry imaging of molecules in biological systems. p 43–72. [Google Scholar]
- Lanni EJ, Dunham SJ, Nemes P, Rubakhin SS, Sweedler JV. 2014. Biomolecular imaging with a C60-SIMS/MALDI dual ion source hybrid mass spectrometer: instrumentation, matrix enhancement, and single cell analysis. J Am Soc Mass Spectrom 25:1897–1907. [DOI] [PubMed] [Google Scholar]
- Lavore A, Perez-Gianmarco L, Esponda-Behrens N, Palacio V, Catalano MI, Rivera-Pomar R, Ons S. 2018. Nezara viridula (Hemiptera: Pentatomidae) transcriptomic analysis and neuropeptidomics. Sci Rep 8:17244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Fan Y, Rubakhin SS, Yoon S, Sweedler JV. 2016. A neuron-in-capillary platform for facile collection and mass spectrometric characterization of a secreted neuropeptide. Sci Rep 6:26940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Atkins N Jr., Hatcher NG, Zamdborg L, Gillette MU, Sweedler JV, Kelleher NL. 2010. Endogenous peptide discovery of the rat circadian clock: a focused study of the suprachiasmatic nucleus by ultrahigh performance tandem mass spectrometry. Mol Cell Proteomics 9:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MR, Rohn MCH, Zanettini C, Coggiano MA, Leggio L, Tanda G. 2019. Effect of systemically administered oxytocin on dose response for methylphenidate self-administration and mesolimbic dopamine levels. Ann N Y Acad Sci 1455:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MR, Scheidweiler KB, Diao XX, Akhlaghi F, Cummins A, Huestis MA, Leggio L, Averbeck BB. 2018. Oxytocin by intranasal and intravenous routes reaches the cerebrospinal fluid in rhesus macaques: determination using a novel oxytocin assay. Mol Psychiatry 23:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinweber BD, Tsaprailis G, Monks TJ, Lau SS. 2009. Improved MALDI-TOF imaging yields increased protein signals at high molecular mass. J Am Soc Mass Spectrom 20:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. 2007. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res 6:1295–1305. [DOI] [PubMed] [Google Scholar]
- Lemaire R, Wisztorski M, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. 2006. MALDI-MS direct tissue analysis of proteins: Improving signal sensitivity using organic treatments. Anal Chem 78:7145–7153. [DOI] [PubMed] [Google Scholar]
- Lewandowska D, ten Have S, Hodge K, Tillemans V, Lamond AI, Brown JW. 2013. Plant SILAC: stable-isotope labelling with amino acids of arabidopsis seedlings for quantitative proteomics. PLoS One 8:e72207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Delafield DG, Li L. 2020. Improved structural elucidation of peptide isomers and their receptors using advanced ion mobility-mass spectrometry. TrAC Trends in Analytical Chemistry 124. [Google Scholar]
- Li G, Ma F, Cao Q, Zheng Z, DeLaney K, Liu R, Li L. 2019. Nanosecond photochemically promoted click chemistry for enhanced neuropeptide visualization and rapid protein labeling. Nat Commun 10:4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Hummon AB. 2011. Imaging mass spectrometry of three-dimensional cell culture systems. Anal Chem 83:8794–8801. [DOI] [PubMed] [Google Scholar]
- Li KW, Gonzalez-Lozano MA, Koopmans F, Smit AB. 2020a. Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome. Front Mol Neurosci 13:564446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sweedler JV. 2008. Peptides in the Brain: Mass Spectrometry-Based Measurement Approaches and ChallengesAnnu. Rev. Anal. Chem Palo Alto: Annual Reviews. p 451–483. [DOI] [PubMed] [Google Scholar]
- Li N, Zhou Y, Wang J, Niu L, Zhang Q, Sun L, Ding X, Guo X, Xie Z, Zhu N, Zhang M, Chen X, Cai T, Yang F. 2020b. Sequential Precipitation and Delipidation Enables Efficient Enrichment of Low-Molecular Weight Proteins and Peptides from Human Plasma. J Proteome Res 19:3340–3351. [DOI] [PubMed] [Google Scholar]
- Li Q, Zubieta JK, Kennedy RT. 2009. Practical aspects of in vivo detection of neuropeptides by microdialysis coupled off-line to capillary LC with multistage MS. Anal Chem 81:2242–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, McGuinness KN, Crespo A, Zhong W. 2018. Characterization of Disulfide-Linked Peptides Using Tandem Mass Spectrometry Coupled with Automated Data Analysis Software. J Am Soc Mass Spectrom 29:903–912. [DOI] [PubMed] [Google Scholar]
- Liang Z, Schmerberg CM, Li L. 2015. Mass spectrometric measurement of neuropeptide secretion in the crab, Cancer borealis, by in vivo microdialysis. Analyst 140:3803–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liessem S, Ragionieri L, Neupert S, Buschges A, Predel R. 2018. Transcriptomic and Neuropeptidomic Analysis of the Stick Insect, Carausius morosus. J Proteome Res 17:2192–2204. [DOI] [PubMed] [Google Scholar]
- Lietz CB, Chen Z, Yun Son C, Pang X, Cui Q, Li L. 2016. Multiple gas-phase conformations of proline-containing peptides: is it always cis/trans isomerization? Analyst 141:4863–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietz CB, Toneff T, Mosier C, Podvin S, O'Donoghue AJ, Hook V. 2018. Phosphopeptidomics Reveals Differential Phosphorylation States and Novel SxE Phosphosite Motifs of Neuropeptides in Dense Core Secretory Vesicles. J Am Soc Mass Spectrom 29:935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Wei P, Keller C, Orefice NS, Shi Y, Li Z, Huang J, Cui Y, Frost DC, Han S, Cross TL, Rey FE, Li L. 2020. Integrated Label-Free and 10-Plex DiLeu Isobaric Tag Quantitative Methods for Profiling Changes in the Mouse Hypothalamic Neuropeptidome and Proteome: Assessment of the Impact of the Gut Microbiome. Anal Chem 92:14021–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Buchberger AR, DeLaney K, Li Z, Li L. 2019. Multifaceted Mass Spectrometric Investigation of Neuropeptide Changes in Atlantic Blue Crab, Callinectes sapidus, in Response to Low pH Stress. J Proteome Res 18:2759–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao Q, Li L. 2019. Isolation and characterization of glycosylated neuropeptides. Methods Enzymol 626:147–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnat I, Tai HC, Jansson ET, Bai L, Romanova EV, Chen TT, Yu K, Chen SA, Zhang Y, Wang ZY, Liu DD, Weiss KR, Jing J, Sweedler JV. 2016. A d-Amino Acid-Containing Neuropeptide Discovery Funnel. Anal Chem 88:11868–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logerot E, Enjalbal C. 2020. Dissociation Pattern of Sodiated Amide Peptides as a Tool for De Novo Sequencing. J Am Soc Mass Spectrom 31:2328–2337. [DOI] [PubMed] [Google Scholar]
- Lopez-Clavijo AF, Barrow MP, Rabbani N, Thornalley PJ, O'Connor PB. 2012. Determination of types and binding sites of advanced glycation end products for substance P. Anal Chem 84:10568–10575. [DOI] [PubMed] [Google Scholar]
- Lu L, Riley NM, Shortreed MR, Bertozzi CR, Smith LM. 2020. O-Pair Search with MetaMorpheus for O-glycopeptide characterization. Nat Methods 17:1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A, Ragionieri L, Liessem S, Becker M, Deininger SO, Neupert S, Predel R. 2019. Enhanced Coverage of Insect Neuropeptides in Tissue Sections by an Optimized Mass-Spectrometry-Imaging Protocol. Anal Chem 91:1980–1988. [DOI] [PubMed] [Google Scholar]
- Ma B 2015. Novor: real-time peptide de novo sequencing software. J Am Soc Mass Spectrom 26:1885–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Johnson R. 2012. De novo sequencing and homology searching. Mol Cell Proteomics 11:O111 014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Bors EK, Dickinson ES, Kwiatkowski MA, Sousa GL, Henry RP, Smith CM, Towle DW, Christie AE, Li L. 2009. Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen Comp Endocrinol 161:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabrouk OS, Kennedy RT. 2012. Simultaneous oxytocin and arg-vasopressin measurements in microdialysates using capillary liquid chromatography-mass spectrometry. J Neurosci Methods 209:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen TD, Hansen LH, Hintze J, Ye Z, Jebari S, Andersen DB, Joshi HJ, Ju T, Goetze JP, Martin C, Rosenkilde MM, Holst JJ, Kuhre RE, Goth CK, Vakhrushev SY, Schjoldager KT. 2020. An atlas of O-linked glycosylation on peptide hormones reveals diverse biological roles. Nat Commun 11:4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes K, Van Liefferinge J, Viaene J, Van Schoors J, Van Wanseele Y, Bechade G, Chambers EE, Morren H, Michotte Y, Vander Heyden Y, Claereboudt J, Smolders I, Van Eeckhaut A. 2014. Improved sensitivity of the nano ultra-high performance liquid chromatography-tandem mass spectrometric analysis of low-concentrated neuropeptides by reducing aspecific adsorption and optimizing the injection solvent. J Chromatogr A 1360:217–228. [DOI] [PubMed] [Google Scholar]
- Magafa V, Matsoukas MT, Karageorgos V, Dermitzaki E, Exarchakou R, Stylos E, Pardalos M, Margioris AN, Varvounis G, Tzakos AG, Spyroulias GA, Liapakis G. 2019. Novel stable analogues of the neurotensin C-terminal hexapeptide containing unnatural amino acids. Amino Acids 51:1009–1022. [DOI] [PubMed] [Google Scholar]
- Mahajan VK, Desiderio DM. 1978. Mass spectrometry of acetylated, permethylated and reduced oligopeptides. Biochemical and Biophysical Research Communications 82:1104–1110. [DOI] [PubMed] [Google Scholar]
- Marciniak P, Witek W, Szymczak M, Pacholska-Bogalska J, Chowanski S, Kuczer M, Rosinski G. 2020. FMRFamide-Related Peptides Signaling Is Involved in the Regulation of Muscle Contractions in Two Tenebrionid Beetles. Front Physiol 11:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark L, Maasz G, Pirger Z. 2012. High resolution spatial distribution of neuropeptides by MALDI imaging mass spectrometry in the terrestrial snail, Helix pomatia. Acta Biol Hung 63 Suppl 2:113–122. [DOI] [PubMed] [Google Scholar]
- Mast DH, Checco JW, Sweedler JV. 2020. Differential Post-Translational Amino Acid Isomerization Found among Neuropeptides in Aplysia californica. ACS Chem Biol 15:272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurais D, Ernande B, Quazuguel P, Severe A, Huelvan C, Madec L, Mouchel O, Soudant P, Robbens J, Huvet A, Zambonino-Infante J. 2015. Evaluation of the impact of polyethylene microbeads ingestion in European sea bass (Dicentrarchus labrax) larvae. Mar Environ Res 112:78–85. [DOI] [PubMed] [Google Scholar]
- McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. 2012. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem 84:7469–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister RG, Metwally H, Sun Y, Konermann L. 2015. Release of Native-like Gaseous Proteins from Electrospray Droplets via the Charged Residue Mechanism: Insights from Molecular Dynamics Simulations. J Am Chem Soc 137:12667–12676. [DOI] [PubMed] [Google Scholar]
- Medzihradszky KF, Chalkley RJ. 2015. Lessons in de novo peptide sequencing by tandem mass spectrometry. Mass Spectrom Rev 34:43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menschaert G, Vandekerckhove TT, Baggerman G, Landuyt B, Sweedler JV, Schoofs L, Luyten W, Van Criekinge W. 2010. A hybrid, de novo based, genome-wide database search approach applied to the sea urchin neuropeptidome. J Proteome Res 9:990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriaux C, Arafah K, Tasiemski A, Wisztorski M, Bruand J, Boidin-Wichlacz C, Desmons A, Debois D, Laprevote O, Brunelle A, Gaasterland T, Macagno E, Fournier I, Salzet M. 2011. Multiple changes in peptide and lipid expression associated with regeneration in the nervous system of the medicinal leech. PLoS One 6:e18359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EG, Izzi-Engbeaya C, Abbara A, Comninos AN, Dhillo WS. 2020. Functions of galanin, spexin and kisspeptin in metabolism, mood and behaviour. Nat Rev Endocrinol. [DOI] [PubMed] [Google Scholar]
- Monroe EB, Annangudi SP, Hatcher NG, Gutstein HB, Rubakhin SS, Sweedler JV. 2008. SIMS and MALDI MS imaging of the spinal cord. Proteomics 8:3746–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe EB, Annangudi SP, Wadhams AA, Richmond TA, Yang N, Southey BR, Romanova EV, Schoofs L, Baggerman G, Sweedler JV. 2018. Exploring the Sea Urchin Neuropeptide Landscape by Mass Spectrometry. J Am Soc Mass Spectrom 29:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe EB, Koszczuk BA, Losh JL, Jurchen JC, Sweedler JV. 2007. Measuring salty samples without adducts with MALDI MS. Int. J. Mass Spectrom 260:237–242. [Google Scholar]
- Muntel J, Kirkpatrick J, Bruderer R, Huang T, Vitek O, Ori A, Reiter L. 2019. Comparison of Protein Quantification in a Complex Background by DIA and TMT Workflows with Fixed Instrument Time. J Proteome Res 18:1340–1351. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Nirasawa T, Takubo T. 2014. Quantitative mass barcode-like image of nicotine in single longitudinally sliced hair sections from long-term smokers by matrix-assisted laser desorption time-of-flight mass spectrometry imaging. J Anal Toxicol 38:349–353. [DOI] [PubMed] [Google Scholar]
- Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. 2011. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics 11:535–553. [DOI] [PubMed] [Google Scholar]
- Neumann ID, Landgraf R. 2012. Balance of brain oxytocin and vasopressin: implications for anxiety, depression, and social behaviors. Trends Neurosci 35:649–659. [DOI] [PubMed] [Google Scholar]
- Neupert S 2018. Single Cell Peptidomics: Approach for Peptide Identification by N-Terminal Peptide DerivatizationPeptidomics. p 369–378. [DOI] [PubMed] [Google Scholar]
- Neupert S, Fusca D, Kloppenburg P, Predel R. 2018. Analysis of Single Neurons by Perforated Patch Clamp Recordings and MALDI-TOF Mass Spectrometry. ACS Chem Neurosci 9:2089–2096. [DOI] [PubMed] [Google Scholar]
- Nielsen BV, Abaye DA. 2013. Influence of electrolytes and a supercharging reagent on charge state distribution and response of neuropeptide ions generated during positive electrospray ionisation mass spectrometry. Eur J Mass Spectrom (Chichester) 19:335–344. [DOI] [PubMed] [Google Scholar]
- Nusbaum MP, Blitz DM, Marder E. 2017. Functional consequences of neuropeptide and small-molecule co-transmission. Nature Reviews Neuroscience 18:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofer D, Linial M. 2014. NeuroPID: a predictor for identifying neuropeptide precursors from metazoan proteomes. Bioinformatics 30:931–940. [DOI] [PubMed] [Google Scholar]
- Ogrinc Potocnik N, Fisher GL, Prop A, Heeren RMA. 2017. Sequencing and Identification of Endogenous Neuropeptides with Matrix-Enhanced Secondary Ion Mass Spectrometry Tandem Mass Spectrometry. Anal Chem 89:8223–8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrinc Potocnik N, Porta T, Becker M, Heeren RM, Ellis SR. 2015. Use of advantageous, volatile matrices enabled by next-generation high-speed matrix-assisted laser desorption/ionization time-of-flight imaging employing a scanning laser beam. Rapid Commun Mass Spectrom 29:2195–2203. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1:376–386. [DOI] [PubMed] [Google Scholar]
- Onorato JM, Xu C, Chen XQ, Rose AV, Generaux C, Lentz K, Shipkova P, Arthur S, Hennan JK, Haskell R, Myers MC, Lawrence RM, Finlay HJ, Basso M, Bostwick J, Fernando G, Garcia R, Hellings S, Hsu MY, Zhang R, Zhao L, Gargalovic P. 2019. Linking (Pyr)(1)apelin-13 pharmacokinetics to efficacy: Stabilization and measurement of a high clearance peptide in rodents. Anal Biochem 568:41–50. [DOI] [PubMed] [Google Scholar]
- OuYang C, Chen B, Li L. 2015. High Throughput In Situ DDA Analysis of Neuropeptides by Coupling Novel Multiplex Mass Spectrometric Imaging (MSI) with Gas-Phase Fractionation. J Am Soc Mass Spectrom 26:1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OuYang C, Liang Z, Li L. 2015. Mass spectrometric analysis of spatio-temporal dynamics of crustacean neuropeptides. Biochim Biophys Acta 1854:798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozalp A, Barroso B, Meijer J, van den Beld C. 2018. Determination of methionine-enkephalin and leucine-enkephalin by LC-MS in human plasma: Study of pre-analytical stability. Anal Biochem 559:24–29. [DOI] [PubMed] [Google Scholar]
- Pailleux F, Beaudry F. 2014. Evaluation of multiple reaction monitoring cubed for the analysis of tachykinin related peptides in rat spinal cord using a hybrid triple quadrupole-linear ion trap mass spectrometer. J Chromatogr B Analyt Technol Biomed Life Sci 947–948:164–167. [DOI] [PubMed] [Google Scholar]
- Paine MRL, Ellis SR, Maloney D, Heeren RMA, Verhaert P. 2018. Digestion-Free Analysis of Peptides from 30-year-old Formalin-Fixed, Paraffin-Embedded Tissue by Mass Spectrometry Imaging. Anal Chem 90:9272–9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos G, Svendsen A, Boyarkin OV, Rizzo TR. 2012. Conformational distribution of bradykinin [bk + 2 H]2+ revealed by cold ion spectroscopy coupled with FAIMS. J Am Soc Mass Spectrom 23:1173–1181. [DOI] [PubMed] [Google Scholar]
- Parker BL, Yang G, Humphrey SJ, Chaudhuri R, Ma X, Peterman S, James DE. 2015. Targeted phosphoproteomics of insulin signaling using data-independent acquisition mass spectrometry. Sci Signal 8:rs6. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. [DOI] [PubMed] [Google Scholar]
- Petruzziello F, Falasca S, Andren PE, Rainer G, Zhang X. 2013. Chronic nicotine treatment impacts the regulation of opioid and non-opioid peptides in the rat dorsal striatum. Mol Cell Proteomics 12:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzziello F, Fouillen L, Wadensten H, Kretz R, Andren PE, Rainer G, Zhang X. 2012. Extensive characterization of Tupaia belangeri neuropeptidome using an integrated mass spectrometric approach. J Proteome Res 11:886–896. [DOI] [PubMed] [Google Scholar]
- Pierson NA, Chen L, Russell DH, Clemmer DE. 2013. Cis–Trans Isomerizations of Proline Residues Are Key to Bradykinin Conformations. J. Am. Chem. Soc 135:3186–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. 2011. Number of solution states of bradykinin from ion mobility and mass spectrometry measurements. J Am Chem Soc 133:13810–13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson NA, Clemmer DE. 2015. An IMS–IMS threshold method for semi-quantitative determination of activation barriers: Interconversion of proline cis↔ trans forms in triply protonated bradykinin. Int. J. Mass Spectrom 377:646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson NA, Valentine SJ, Clemmer DE. 2010. Evidence for a quasi-equilibrium distribution of states for bradykinin [M + 3H]3+ ions in the gas phase. J Phys Chem B 114:7777–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirman D 2015. Quantitative profiling of tissue drug distribution by MS imaging. Bioanalysis 7:2649–2656. [DOI] [PubMed] [Google Scholar]
- Potts GK, Voigt EA, Bailey DJ, Rose CM, Westphall MS, Hebert AS, Yin J, Coon JJ. 2016. Neucode Labels for Multiplexed, Absolute Protein Quantification. Analytical Chemistry 88:3295–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratavieira M, da Silva Menegasso AR, Garcia AM, Dos Santos DS, Gomes PC, Malaspina O, Palma MS. 2014. MALDI imaging analysis of neuropeptides in the Africanized honeybee (Apis mellifera) brain: effect of ontogeny. J Proteome Res 13:3054–3064. [DOI] [PubMed] [Google Scholar]
- Predel R, Neupert S, Derst C, Reinhardt K, Wegener C. 2018. Neuropeptidomics of the Bed Bug Cimex lectularius. J Proteome Res 17:440–454. [DOI] [PubMed] [Google Scholar]
- Predel R, Neupert S, Garczynski SF, Crim JW, Brown MR, Russell WK, Kahnt J, Russell DH, Nachman RJ. 2010. Neuropeptidomics of the mosquito Aedes aegypti. J Proteome Res 9:2006–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Yang Y, Li N, Cheng T, Wang X, Liu J, Li X, Desiderio DM, Zhan X. 2018. Prolactin Variants in Human Pituitaries and Pituitary Adenomas Identified With Two-Dimensional Gel Electrophoresis and Mass Spectrometry. Front Endocrinol (Lausanne) 9:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistgaard EM, Groftehauge MK, Madsen P, Pallesen LT, Christensen B, Sorensen ES, Nissen P, Petersen CM, Thirup SS. 2014. Revisiting the structure of the Vps10 domain of human sortilin and its interaction with neurotensin. Protein Sci 23:1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragionieri L, Predel R. 2020. The neuropeptidome of Carabus (Coleoptera, Adephaga: Carabidae). Insect Biochem Mol Biol 118:103309. [DOI] [PubMed] [Google Scholar]
- Ranc V, Petruzziello F, Kretz R, Argandona EG, Zhang X, Rainer G. 2012. Broad characterization of endogenous peptides in the tree shrew visual system. J Proteomics 75:2526–2535. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Aoki-Kinoshita KF, Campbell MP, Kawano S, Lutteke T, Okuda S, Shinmachi D, Shikanai T, Sawaki H, Toukach P, Matsubara M, Yamada I, Narimatsu H. 2015. GlycoRDF: an ontology to standardize glycomics data in RDF. Bioinformatics 31:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore D, Aboufazeli F, Dodds ED. 2015. Obtaining complementary polypeptide sequence information from a single precursor ion packet via sequential ion mobility-resolved electron transfer and vibrational activation. Analyst 140:7175–7183. [DOI] [PubMed] [Google Scholar]
- Resetar Maslov D, Svirkova A, Allmaier G, Marchetti-Deschamann M, Kraljevic Pavelic S. 2019. Optimization of MALDI-TOF mass spectrometry imaging for the visualization and comparison of peptide distributions in dry-cured ham muscle fibers. Food Chem 283:275–286. [DOI] [PubMed] [Google Scholar]
- Riley NM, Malaker SA, Driessen MD, Bertozzi CR. 2020. Optimal Dissociation Methods Differ for N- and O-Glycopeptides. J Proteome Res 19:3286–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud G, Garrard KP, Barry JA, Muddiman DC. 2013. MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectrom 24:718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha B, Ruiz-Romero C, Blanco FJ. 2017. Mass spectrometry imaging: a novel technology in rheumatology. Nat Rev Rheumatol 13:52–63. [DOI] [PubMed] [Google Scholar]
- Rodrigues LR, de Oliveira DN, Ferreira MS, Catharino RR. 2014. In situ assessment of atorvastatin impurity using MALDI mass spectrometry imaging (MALDI-MSI). Anal Chim Acta 818:32–38. [DOI] [PubMed] [Google Scholar]
- Romanova EV, Dowd SE, Sweedler JV. 2013. Quantitation of endogenous peptides using mass spectrometry based methods. Curr Opin Chem Biol 17:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova EV, Hatcher NG, Rubakhin SS, Sweedler JV. 2009. Characterizing intercellular signaling peptides in drug addiction. Neuropharmacology 56 Suppl 1:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova EV, Rubakhin SS, Sweedler JV. 2008. One-step sampling, extraction, and storage protocol for peptidomics using dihydroxybenzoic Acid. Anal Chem 80:3379–3386. [DOI] [PubMed] [Google Scholar]
- Romanova EV, Sweedler JV. 2015. Peptidomics for the discovery and characterization of neuropeptides and hormones. Trends Pharmacol Sci 36:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompp A, Spengler B. 2013. Mass spectrometry imaging with high resolution in mass and space. Histochem Cell Biol 139:759–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roushan A, Wilson GM, Kletter D, Sen KI, Tang WH, Kil YJ, Carlson E, Bern M. 2020. Peak Filtering, Peak Annotation, and Wildcard Search for Glycoproteomics. Mol Cell Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin SS, Sweedler JV. 2008. Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal Chem 80:7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi M, Kamali S, Beaudry F. 2019. Neuropeptidomics: Comparison of parallel reaction monitoring and data-independent acquisition for the analysis of neuropeptides using high-resolution mass spectrometry. Biomed Chromatogr 33:e4523. [DOI] [PubMed] [Google Scholar]
- Salem JB, Nkambeu B, Beaudry F. 2018. Characterization of neuropeptide K processing in rat spinal cord S9 fractions using high-resolution quadrupole-Orbitrap mass spectrometry. Biomed Chromatogr 32:e4204. [DOI] [PubMed] [Google Scholar]
- Salisbury JP, Boggio KJ, Hsu YW, Quijada J, Sivachenko A, Gloeckner G, Kowalski PJ, Easterling ML, Rosbash M, Agar JN. 2013. A rapid MALDI-TOF mass spectrometry workflow for Drosophila melanogaster differential neuropeptidomics. Mol Brain 6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargaeva NP, Lin C, O'Connor PB. 2011. Differentiating N-terminal aspartic and isoaspartic acid residues in peptides. Anal Chem 83:6675–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Osaki T, Minamino N. 2013. Large-scale identification of endogenous secretory peptides using electron transfer dissociation mass spectrometry. Mol Cell Proteomics 12:700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarff CA, Thalassinos K, Hilton GR, Scrivens JH. 2008. Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements. Rapid Commun Mass Spectrom 22:3297–3304. [DOI] [PubMed] [Google Scholar]
- Schmerberg CM, Li L. 2013. Mass spectrometric detection of neuropeptides using affinity-enhanced microdialysis with antibody-coated magnetic nanoparticles. Anal Chem 85:915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerberg CM, Liang Z, Li L. 2015. Data-independent MS/MS quantification of neuropeptides for determination of putative feeding-related neurohormones in microdialysate. ACS Chem Neurosci 6:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J, Realis-Doyelle E, Dubos MP, Lefranc B, Leprince J, Favrel P. 2019. Characterization of an evolutionarily conserved calcitonin signalling system in a lophotrochozoan, the Pacific oyster (Crassostrea gigas). J Exp Biol 222. [DOI] [PubMed] [Google Scholar]
- Secher A, Kelstrup CD, Conde-Frieboes KW, Pyke C, Raun K, Wulff BS, Olsen JV. 2016. Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nat Commun 7:11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. 2008. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am Soc Mass Spectrom 19:1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibert C, Sakmar TP. 2008. Toward a framework for sulfoproteomics: Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers 90:459–477. [DOI] [PubMed] [Google Scholar]
- Seidler J, Zinn N, Boehm ME, Lehmann WD. 2010. De novo sequencing of peptides by MS/MS. Proteomics 10:634–649. [DOI] [PubMed] [Google Scholar]
- Servage KA, Silveira JA, Fort KL, Russell DH. 2015. From solution to gas phase: the implications of intramolecular interactions on the evaporative dynamics of substance P during electrospray ionization. J Phys Chem B 119:4693–4698. [DOI] [PubMed] [Google Scholar]
- Servage KA, Silveira JA, Fort KL, Russell DH. 2016. Cryogenic Ion Mobility-Mass Spectrometry: Tracking Ion Structure from Solution to the Gas Phase. Acc Chem Res 49:1421–1428. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Nilsson A, Goodwin RJ, Kallback P, Schintu N, Zhang X, Crossman AR, Bezard E, Svenningsson P, Andren PE. 2014. Direct targeted quantitative molecular imaging of neurotransmitters in brain tissue sections. Neuron 84:697–707. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Nilsson A, Kallback P, Karlsson O, Zhang X, Svenningsson P, Andren PE. 2015. Pyrylium Salts as Reactive Matrices for MALDI-MS Imaging of Biologically Active Primary Amines. J Am Soc Mass Spectrom 26:934–939. [DOI] [PubMed] [Google Scholar]
- Shariatgorji M, Svenningsson P, Andren PE. 2014. Mass spectrometry imaging, an emerging technology in neuropsychopharmacology. Neuropsychopharmacology 39:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Pierson NA, Valentine SJ, Clemmer DE. 2012. Conformation types of ubiquitin [M+8H]8+ Ions from water:methanol solutions: evidence for the N and A States in aqueous solution. J Phys Chem B 116:3344–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Holliday AE, Bohrer BC, Kim D, Servage KA, Russell DH, Clemmer DE. 2016. "Wet" Versus "Dry" Folding of Polyproline. J Am Soc Mass Spectrom 27:1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Holliday AE, Shi H, Zhu F, Ewing MA, Russell DH, Clemmer DE. 2014. Characterizing intermediates along the transition from polyproline I to polyproline II using ion mobility spectrometry-mass spectrometry. J. Am. Chem. Soc 136:12702–12711. [DOI] [PubMed] [Google Scholar]
- Silva EMP, Varandas P, Melo T, Barros C, Alencastre IS, Barreiros L, Domingues P, Lamghari M, Domingues MRM, Segundo MA. 2018. Gas-phase structural characterization of neuropeptides Y Y1 receptor antagonists using mass spectrometry: Orbitrap vs triple quadrupole. J Pharm Biomed Anal 151:227–234. [DOI] [PubMed] [Google Scholar]
- Silveira JA, Fort KL, Kim D, Servage KA, Pierson NA, Clemmer DE, Russell DH. 2013a. From solution to the gas phase: stepwise dehydration and kinetic trapping of substance P reveals the origin of peptide conformations. J Am Chem Soc 135:19147–19153. [DOI] [PubMed] [Google Scholar]
- Silveira JA, Servage KA, Gamage CM, Russell DH. 2013b. Cryogenic ion mobility-mass spectrometry captures hydrated ions produced during electrospray ionization. J Phys Chem A 117:953–961. [DOI] [PubMed] [Google Scholar]
- Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE. 2007. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2-20 and peptides as sample quality indicators. Proteomics 7:4445–4456. [DOI] [PubMed] [Google Scholar]
- Smith D, Griffin JF. 1978. Conformation of [Leu5] enkephalin from X-ray diffraction: features important for recognition at opiate receptor. Science 199:1214–1216. [DOI] [PubMed] [Google Scholar]
- Sobott F, Watt SJ, Smith J, Edelmann MJ, Kramer HB, Kessler BM. 2009. Comparison of CID versus ETD based MS/MS fragmentation for the analysis of protein ubiquitination. J Am Soc Mass Spectrom 20:1652–1659. [DOI] [PubMed] [Google Scholar]
- Song Y, Liu YM. 2008. Quantitation of cardioexcitatory Asn-D-Trp-Phe-NH2 diastereomers in Aplysia's central nervous system by nanoscale liquid chromatography-tandem mass spectrometry. J Mass Spectrom 43:1285–1290. [DOI] [PubMed] [Google Scholar]
- Sosnowski P, Zera T, Wilenska B, Szczepanska-Sadowska E, Misicka A. 2015. Imaging and identification of endogenous peptides from rat pituitary embedded in egg yolk. Rapid Commun Mass Spectrom 29:327–335. [DOI] [PubMed] [Google Scholar]
- Southey BR, Lee JE, Zamdborg L, Atkins N Jr., Mitchell JW, Li M, Gillette MU, Kelleher NL, Sweedler JV. 2014. Comparing label-free quantitative peptidomics approaches to characterize diurnal variation of peptides in the rat suprachiasmatic nucleus. Anal Chem 86:443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Moreira L, Campos-Salinas J, Caro M, Gonzalez-Rey E. 2011. Neuropeptides as Pleiotropic Modulators of the Immune Response. Neuroendocrinology 94:89–100. [DOI] [PubMed] [Google Scholar]
- Soyez D, Toullec JY, Montagne N, Ollivaux C. 2011. Experimental strategies for the analysis of D-amino acid containing peptides in crustaceans: a review. J Chromatogr B Analyt Technol Biomed Life Sci 879:3102–3107. [DOI] [PubMed] [Google Scholar]
- Stauber J, Lemaire R, Franck J, Bonnel D, Croix D, Day R, Wisztorski M, Fournier I, Salzet M. 2008. MALDI imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J Proteome Res 7:969–978. [DOI] [PubMed] [Google Scholar]
- Steinhoff MS, von Mentzer B, Geppetti P, Pothoulakis C, Bunnett NW. 2014. Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiological reviews 94:265–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmler EA, Barton EE, Esonu OK, Polasky DA, Onderko LL, Bergeron AB, Christie AE, Dickinson PS. 2013. C-terminal methylation of truncated neuropeptides: an enzyme-assisted extraction artifact involving methanol. Peptides 46:108–125. [DOI] [PubMed] [Google Scholar]
- Sterkel M, Urlaub H, Rivera-Pomar R, Ons S. 2011. Functional proteomics of neuropeptidome dynamics during the feeding process of Rhodnius prolixus. J Proteome Res 10:3363–3371. [DOI] [PubMed] [Google Scholar]
- Stocks BB, Melanson JE. 2018. In-Source Reduction of Disulfide-Bonded Peptides Monitored by Ion Mobility Mass Spectrometry. J Am Soc Mass Spectrom 29:742–751. [DOI] [PubMed] [Google Scholar]
- Stocks BB, Melanson JE. 2019. Corona discharge electrospray ionization of formate-containing solutions enables in-source reduction of disulfide bonds. Anal Bioanal Chem 411:4729–4737. [DOI] [PubMed] [Google Scholar]
- Strohalm M, Strohalm J, Kaftan F, Krasny L, Volny M, Novak P, Ulbrich K, Havlicek V. 2011. Poly[N-(2-hydroxypropyl)methacrylamide]-based tissue-embedding medium compatible with MALDI mass spectrometry imaging experiments. Anal Chem 83:5458–5462. [DOI] [PubMed] [Google Scholar]
- Studneva I, Serebryakova L, Veselova O, Pal'keeva M, Molokoedov A, Ovchinnikov M, Konovalova G, Lankin V, Sidorova M, Pisarenko O. 2019. Galanin receptors activation modulates myocardial metabolic and antioxidant responses to ischaemia/reperfusion stress. Clin Exp Pharmacol Physiol 46:1174–1182. [DOI] [PubMed] [Google Scholar]
- Sturm RM, Dowell JA, Li L. 2010. Rat brain neuropeptidomics: tissue collection, protease inhibition, neuropeptide extraction, and mass spectrometric analysis. Methods Mol Biol 615:217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm RM, Greer T, Chen R, Hensen B, Li L. 2013a. Comparison of NIMS and MALDI platforms for neuropeptide and lipid mass spectrometric imaging in C. borealis brain tissue. Anal Methods 5:1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm RM, Greer T, Woodards N, Gemperline E, Li L. 2013b. Mass spectrometric evaluation of neuropeptidomic profiles upon heat stabilization treatment of neuroendocrine tissues in crustaceans. J Proteome Res 12:743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm S, Predel R. 2014. Serine phosphorylation of CAPA pyrokinin in cockroaches-a taxon-specific posttranslational modification. Peptides 57:52–58. [DOI] [PubMed] [Google Scholar]
- Sun N, Walch A. 2013. Qualitative and quantitative mass spectrometry imaging of drugs and metabolites in tissue at therapeutic levels. Histochem Cell Biol 140:93–104. [DOI] [PubMed] [Google Scholar]
- Svensson M, Boren M, Skold K, Falth M, Sjogren B, Andersson M, Svenningsson P, Andren PE. 2009. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res 8:974–981. [DOI] [PubMed] [Google Scholar]
- Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. 2004. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A 101:9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Sato N, Ikimura K, Nishino H, Rakugi H, Morishita R. 2011. Novel microdialysis method to assess neuropeptides and large molecules in free-moving mouse. Neuroscience 186:110–119. [DOI] [PubMed] [Google Scholar]
- Tang Y, Li R, Lin G, Li L. 2014. PEP search in MyCompoundID: detection and identification of dipeptides and tripeptides using dimethyl labeling and hydrophilic interaction liquid chromatography tandem mass spectrometry. Anal Chem 86:3568–3574. [DOI] [PubMed] [Google Scholar]
- Tashima AK, Fricker LD. 2018. Quantitative Peptidomics with Five-plex Reductive Methylation labels. J Am Soc Mass Spectrom 29:866–878. [DOI] [PubMed] [Google Scholar]
- Taverna D, Norris JL, Caprioli RM. 2015. Histology-directed microwave assisted enzymatic protein digestion for MALDI MS analysis of mammalian tissue. Anal Chem 87:670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegge AN, Southey BR, Sweedler JV, Rodriguez-Zas SL. 2008. Comparative analysis of neuropeptide cleavage sites in human, mouse, rat, and cattle. Mamm Genome 19:106–120. [DOI] [PubMed] [Google Scholar]
- Tiemeyer M, Aoki K, Paulson J, Cummings RD, York WS, Karlsson NG, Lisacek F, Packer NH, Campbell MP, Aoki NP, Fujita A, Matsubara M, Shinmachi D, Tsuchiya S, Yamada I, Pierce M, Ranzinger R, Narimatsu H, Aoki-Kinoshita KF. 2017. GlyTouCan: an accessible glycan structure repository. Glycobiology 27:915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmaand EG, Anapindi KDB, De La Toba EA, Guo CJ, Krebs J, Lenhart AE, Liu Q, Sweedler JV. 2020. Quantitative Characterization of the Neuropeptide Level Changes in Dorsal Horn and Dorsal Root Ganglia Regions of the Murine Itch Models. J Proteome Res 19:1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NH, Qiao R, Xin L, Chen X, Liu C, Zhang X, Shan B, Ghodsi A, Li M. 2019. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat Methods 16:63–66. [DOI] [PubMed] [Google Scholar]
- Tran NH, Zhang X, Xin L, Shan B, Li M. 2017. De novo peptide sequencing by deep learning. Proc Natl Acad Sci U S A 114:8247–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede D, Schiffler S, Becker M, Wirtz S, Steinhorst K, Strehlow J, Aichler M, Kobarg JH, Oetjen J, Dyatlov A, Heldmann S, Walch A, Thiele H, Maass P, Alexandrov T. 2012. Exploring three-dimensional matrix-assisted laser desorption/ionization imaging mass spectrometry data: three-dimensional spatial segmentation of mouse kidney. Anal Chem 84:6079–6087. [DOI] [PubMed] [Google Scholar]
- Trimpin S, Inutan ED, Herath TN, McEwen CN. 2010. Matrix-assisted laser desorption/ionization mass spectrometry method for selectively producing either singly or multiply charged molecular ions. Anal Chem 82:11–15. [DOI] [PubMed] [Google Scholar]
- Trimpin S, Ren Y, Wang B, Lietz CB, Richards AL, Marshall DD, Inutan ED. 2011. Extending the laserspray ionization concept to produce highly charged ions at high vacuum on a time-of-flight mass analyzer. Anal Chem 83:5469–5475. [DOI] [PubMed] [Google Scholar]
- Tsai PL, Chen SF. 2017. A Brief Review of Bioinformatics Tools for Glycosylation Analysis by Mass Spectrometry. Mass Spectrom (Tokyo) 6:S0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C, Li J, Shen S, Sheng Q, Shyr Y, Qu J. 2016. Performance Investigation of Proteomic Identification by HCD/CID Fragmentations in Combination with High/Low-Resolution Detectors on a Tribrid, High-Field Orbitrap Instrument. PLoS One 11:e0160160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bael S, Edwards SL, Husson SJ, Temmerman L. 2018a. Identification of Endogenous Neuropeptides in the Nematode C. elegans Using Mass Spectrometry. Methods Mol Biol 1719:271–291. [DOI] [PubMed] [Google Scholar]
- Van Bael S, Watteyne J, Boonen K, De Haes W, Menschaert G, Ringstad N, Horvitz HR, Schoofs L, Husson SJ, Temmerman L. 2018b. Mass spectrometric evidence for neuropeptide-amidating enzymes in Caenorhabditis elegans. J Biol Chem 293:6052–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijck A, Hayakawa E, Landuyt B, Baggerman G, Van Dam D, Luyten W, Schoofs L, De Deyn PP. 2011. Comparison of extraction methods for peptidomics analysis of mouse brain tissue. J Neurosci Methods 197:231–237. [DOI] [PubMed] [Google Scholar]
- van Remoortere A, van Zeijl RJ, van den Oever N, Franck J, Longuespee R, Wisztorski M, Salzet M, Deelder AM, Fournier I, McDonnell LA. 2010. MALDI imaging and profiling MS of higher mass proteins from tissue. J Am Soc Mass Spectrom 21:1922–1929. [DOI] [PubMed] [Google Scholar]
- Van Wanseele Y, Viaene J, Van den Borre L, Dewachter K, Vander Heyden Y, Smolders I, Van Eeckhaut A. 2017. LC-method development for the quantification of neuromedin-like peptides. Emphasis on column choice and mobile phase composition. J Pharm Biomed Anal 137:104–112. [DOI] [PubMed] [Google Scholar]
- Verhaert PD, Conaway MCP, Pekar TM, Miller K. 2007. Neuropeptide imaging on an LTQ with vMALDI source: The complete ‘all-in-one’ peptidome analysis. International Journal of Mass Spectrometry 260:177–184. [Google Scholar]
- Verhaert PDEM, Pinkse MWH, Strupat K, Conaway MCP. 2010. Imaging of similar mass neuropeptides in neuronal tissue by enhanced resolution MALDI MS with an ion trap - Orbitrap hybrid instrument. Methods Mol. Biol. (N. Y., NY, U. S.) 656:433–449. [DOI] [PubMed] [Google Scholar]
- Vocat C, Dunand M, Hubers SA, Bourdillon N, Millet GP, Brown NJ, Wuerzner G, Grouzmann E, Eugster PJ. 2020. Quantification of Neuropeptide Y and Four of Its Metabolites in Human Plasma by Micro-UHPLC-MS/MS. Anal Chem 92:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bohlen und Halbach O. 2005. The renin-angiotensin system in the mammalian central nervous system. Curr Protein Pept Sci 6:355–371. [DOI] [PubMed] [Google Scholar]
- Voronina L, Rizzo TR. 2015. Spectroscopic studies of kinetically trapped conformations in the gas phase: the case of triply protonated bradykinin. Phys Chem Chem Phys 17:25828–25836. [DOI] [PubMed] [Google Scholar]
- Vrkoslav V, Muck A, Brown JM, Hubalek M, Cvacka J. 2018. The matrix-assisted laser desorption/ionisation in-source decay of peptides using ion mobility enabled quadrupole-time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 32:2099–2105. [DOI] [PubMed] [Google Scholar]
- Vu NQ, Buchberger AR, Johnson J, Li L. 2021. Complementary neuropeptide detection in crustacean brain by mass spectrometry imaging using formalin and alternative aqueous tissue washes. Analytical and Bioanalytical Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu NQ, DeLaney K, Li L. 2020. Neuropeptidomics: Improvements in Mass Spectrometry Imaging Analysis and Recent Advancements. Curr Protein Pept Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Fang S, Wohlhueter RM. 2009. N-terminal derivatization of peptides with isothiocyanate analogues promoting Edman-type cleavage and enhancing sensitivity in electrospray ionization tandem mass spectrometry analysis. Anal Chem 81:1893–1900. [DOI] [PubMed] [Google Scholar]
- Wang H, Chung-Davidson YW, Li W. 2014. Identification and quantification of sea lamprey gonadotropin-releasing hormones by electrospray ionization tandem mass spectrometry. J Chromatogr A 1345:98–106. [DOI] [PubMed] [Google Scholar]
- Wang H, DeGnore JP, Kelly BD, True J, Garsha K, Bieniarz C. 2015a. A technique for relative quantitation of cancer biomarkers in formalin-fixed, paraffin-embedded (FFPE) tissue using stable-isotope-label based mass spectrometry imaging (SILMSI). J Mass Spectrom 50:1088–1095. [DOI] [PubMed] [Google Scholar]
- Wang J, Ye H, Zhang Z, Xiang F, Girdaukas G, Li L. 2011. Advancing matrix-assisted laser desorption/ionization-mass spectrometric imaging for capillary electrophoresis analysis of peptides. Anal Chem 83:3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Marti DW, Anderson RE. 2019. Development and Validation of a Simple LC-MS Method for the Quantification of Oxytocin in Dog Saliva. Molecules 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xiao C, Li Y, Ling L, Chen X, Guo X. 2017. A Surface Pattern on MALDI Steel Plate for One-Step In-Situ Self-Desalting and Enrichment of Peptides/Proteins. J Am Soc Mass Spectrom 28:428–433. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu J, Kang Q. 2021. Neuromodulation of bone: Role of different peptides and their interactions (Review). Mol Med Rep 23:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang M, Yin S, Jang R, Wang J, Xue Z, Xu T. 2015b. NeuroPep: a comprehensive resource of neuropeptides. Database (Oxford) 2015:bav038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardman JH, Zhang X, Gagnon S, Castro LM, Zhu X, Steiner DF, Day R, Fricker LD. 2010. Analysis of peptides in prohormone convertase 1/3 null mouse brain using quantitative peptidomics. J Neurochem 114:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnke S, von Helden G, Pagel K. 2015. Analyzing the higher order structure of proteins with conformer-selective ultraviolet photodissociation. Proteomics 15:2804–2812. [DOI] [PubMed] [Google Scholar]
- Wei Y, Lin D, Xu Z, Gao X, Zeng C, Ye H. 2020. A Possible Role of Crustacean Cardioactive Peptide in Regulating Immune Response in Hepatopancreas of Mud Crab. Front Immunol 11:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesson DE, Jo CH, Simoni J. 2015. Angiotensin II-mediated GFR decline in subtotal nephrectomy is due to acid retention associated with reduced GFR. Nephrol Dial Transplant 30:762–770. [DOI] [PubMed] [Google Scholar]
- Willie JT, Lim MM, Bennett RE, Azarion AA, Schwetye KE, Brody DL. 2012. Controlled cortical impact traumatic brain injury acutely disrupts wakefulness and extracellular orexin dynamics as determined by intracerebral microdialysis in mice. J Neurotrauma 29:1908–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RE, Jaquins-Gerstl A, Weber SG. 2018. On-Column Dimethylation with Capillary Liquid Chromatography-Tandem Mass Spectrometry for Online Determination of Neuropeptides in Rat Brain Microdialysate. Anal Chem 90:4561–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. 2008. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc Natl Acad Sci U S A 105:18120–18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiwatpanit T, Powers B, Dickinson PS. 2012. Inter-animal variability in the effects of C-type allatostatin on the cardiac neuromuscular system in the lobster Homarus americanus. J Exp Biol 215:2308–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin CL, Maxon M, Desaire H. 2013. Software for automated interpretation of mass spectrometry data from glycans and glycopeptides. Analyst 138:2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Monroe ME, Xu Z, Slysz GW, Payne SH, Rodland KD, Liu T, Smith RD. 2015a. An Optimized Informatics Pipeline for Mass Spectrometry-Based Peptidomics. J Am Soc Mass Spectrom 26:2002–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XN, Zhang T, Qian NS, Guo XD, Yang HJ, Huang KB, Luo GQ, Xiang W, Deng WT, Dai GH, Peng KR, Pan SY. 2015b. Antinociceptive effects of endomorphin-2: suppression of substance P release in the inflammatory pain model rat. Neurochem Int 82:1–9. [DOI] [PubMed] [Google Scholar]
- Wyttenbach T, Grabenauer M, Thalassinos K, Scrivens JH, Bowers MT. 2009. The effect of calcium ions and peptide ligands on the relative stabilities of the calmodulin dumbbell and compact structures. J. Phys. Chem. B 114:437–447. [DOI] [PubMed] [Google Scholar]
- Xiang F, Ye H, Chen R, Fu Q, Li L. 2010. N,N-dimethyl leucines as novel isobaric tandem mass tags for quantitative proteomics and peptidomics. Anal Chem 82:2817–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Romanova EV, Sweedler JV. 2011. Neuropeptidomics of the Mammalian Brain. In: Li KW, Editor Neuroproteomics. Totowa: Humana Press Inc. p 229–242. [Google Scholar]
- Xie K, Fidler IJ. 1998. Therapy of cancer metastasis by activation of the inducible nitric oxide synthase. Cancer Metastasis Rev 17:55–75. [DOI] [PubMed] [Google Scholar]
- Xu H, Shi X, Li X, Zou J, Zhou C, Liu W, Shao H, Chen H, Shi L. 2020. Neurotransmitter and neuropeptide regulation of mast cell function: a systematic review. Journal of Neuroinflammation 17:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Desiderio DM. 1982. Measurement of endogenous leucine enkephalin in canine caudate nuclei and hypothalami with high-performance liquid chromatography and field-desorption mass spectrometry. Analytical Biochemistry 127:213–221. [DOI] [PubMed] [Google Scholar]
- Yanes O, Woo HK, Northen TR, Oppenheimer SR, Shriver L, Apon J, Estrada MN, Potchoiba MJ, Steenwyk R, Manchester M, Siuzdak G. 2009. Nanostructure initiator mass spectrometry: tissue imaging and direct biofluid analysis. Anal Chem 81:2969–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Anapindi KDB, Romanova EV, Rubakhin SS, Sweedler JV. 2017. Improved identification and quantitation of mature endogenous peptides in the rodent hypothalamus using a rapid conductive sample heating system. Analyst 142:4476–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Anapindi KDB, Rubakhin SS, Wei P, Yu Q, Li L, Kenny PJ, Sweedler JV. 2018. Neuropeptidomics of the Rat Habenular Nuclei. J Proteome Res 17:1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda A, Tatsu Y, Kawata Y, Akizawa T, Shigeri Y. 2011. Post-translational modifications of pro-opiomelanocrtin related hormones in medaka pituitary based on mass spectrometric analyses. Peptides 32:2127–2130. [DOI] [PubMed] [Google Scholar]
- Ye H, Greer T, Li L. 2012. Probing neuropeptide signaling at the organ and cellular domains via imaging mass spectrometry. J Proteomics 75:5014–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Hui L, Kellersberger K, Li L. 2013. Mapping of neuropeptides in the crustacean stomatogastric nervous system by imaging mass spectrometry. J Am Soc Mass Spectrom 24:134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Wang J, Tian Z, Ma F, Dowell J, Bremer Q, Lu G, Baldo B, Li L. 2017. Quantitative Mass Spectrometry Reveals Food Intake-Induced Neuropeptide Level Changes in Rat Brain: Functional Assessment of Selected Neuropeptides as Feeding Regulators. Mol Cell Proteomics 16:1922–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Wang J, Zhang Z, Jia C, Schmerberg C, Catherman AD, Thomas PM, Kelleher NL, Li L. 2015. Defining the Neuropeptidome of the Spiny Lobster Panulirus interruptus Brain Using a Multidimensional Mass Spectrometry-Based Platform. J Proteome Res 14:4776–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh JGC, Pandit AA, Zandawala M, Nassel DR, Davies SA, Dow JAT. 2017. DINeR: Database for Insect Neuropeptide Research. Insect Biochem Mol Biol 86:9–19. [DOI] [PubMed] [Google Scholar]
- Yew JY, Kutz KK, Dikler S, Messinger L, Li L, Stretton AO. 2005. Mass spectrometric map of neuropeptide expression in Ascaris suum. J Comp Neurol 488:396–413. [DOI] [PubMed] [Google Scholar]
- Yew JY, Wang Y, Barteneva N, Dikler S, Kutz-Naber KK, Li L, Kravitz EA. 2009. Analysis of neuropeptide expression and localization in adult drosophila melanogaster central nervous system by affinity cell-capture mass spectrometry. J Proteome Res 8:1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Hou XW, Romanova EV, Sweedler JV. 2011. Neuropeptidomics: Mass Spectrometry-Based Qualitative and Quantitative Analysis. In: Merighi A, Editor Neuropeptides: Methods and Protocols. Totowa: Humana Press Inc. p 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Park S, Kim MS, Lee CY. 2018. Concomitant desalting and concentration of neuropeptides on a donut-shaped surface pattern for MALDI mass spectrometry. Chem Commun 54:5688–5691. [DOI] [PubMed] [Google Scholar]
- Yu Q, Canales A, Glover MS, Das R, Shi X, Liu Y, Keller MP, Attie AD, Li L. 2017. Targeted Mass Spectrometry Approach Enabled Discovery of O-Glycosylated Insulin and Related Signaling Peptides in Mouse and Human Pancreatic Islets. Anal Chem 89:9184–9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Liang Z, OuYang C, Li L. 2015a. Biologically Active Peptides in Invertebrates. Colloquium Series on Neuropeptides 3:1–76. [Google Scholar]
- Yu Q, OuYang C, Liang Z, Li L. 2014. Mass spectrometric characterization of the crustacean neuropeptidome. EuPA Open Proteomics 3:152–170. [Google Scholar]
- Yu X, Khani A, Ye X, Petruzziello F, Gao H, Zhang X, Rainer G. 2015b. High-Efficiency Recognition and Identification of Disulfide Bonded Peptides in Rat Neuropeptidome Using Targeted Electron Transfer Dissociation Tandem Mass Spectrometry. Anal Chem 87:11646–11651. [DOI] [PubMed] [Google Scholar]
- Zanivan S, Krueger M, Mann M. 2011. In Vivo Quantitative Proteomics: The SILAC Mouse. In: Shimaoka M, Editor Integrin and Cell Adhesion Molecules: Methods and Protocols. Totowa: Humana Press Inc. p 435–450. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ge W, Ruan G, Cai X, Guo T. 2020a. Data-Independent Acquisition Mass Spectrometry-Based Proteomics and Software Tools: A Glimpse in 2020. Proteomics 20:e1900276. [DOI] [PubMed] [Google Scholar]
- Zhang L, Khattar N, Kemenes I, Kemenes G, Zrinyi Z, Pirger Z, Vertes A. 2018a. Subcellular Peptide Localization in Single Identified Neurons by Capillary Microsampling Mass Spectrometry. Sci Rep 8:12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Y, Li Y, Li W, Li R, Xie X, Wang S, Hu X, Zhang L, Bao Z. 2018b. Identification and Characterization of Neuropeptides by Transcriptome and Proteome Analyses in a Bivalve Mollusc Patinopecten yessoensis. Front Genet 9:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Petruzziello F, Zani F, Fouillen L, Andren PE, Solinas G, Rainer G. 2012a. High identification rates of endogenous neuropeptides from mouse brain. J Proteome Res 11:2819–2827. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Buchberger A, Muthuvel G, Li L. 2015. Expression and distribution of neuropeptides in the nervous system of the crab Carcinus maenas and their roles in environmental stress. Proteomics 15:3969–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, DeLaney K, Hui L, Wang J, Sturm RM, Li L. 2018c. A Multifaceted Mass Spectrometric Method to Probe Feeding Related Neuropeptide Changes in Callinectes sapidus and Carcinus maenas. J Am Soc Mass Spectrom 29:948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yanez Guerra LA, Egertova M, Zampronio CG, Jones AM, Elphick MR. 2020b. Molecular and functional characterization of somatostatin-type signalling in a deuterostome invertebrate. Open Biol 10:200172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jia C, Li L. 2012. Neuropeptide analysis with liquid chromatography-capillary electrophoresis-mass spectrometric imaging. J Sep Sci 35:1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kuang J, Li L. 2013. Liquid chromatography-matrix-assisted laser desorption/ionization mass spectrometric imaging with sprayed matrix for improved sensitivity, reproducibility and quantitation. Analyst 138:6600–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ye H, Wang J, Hui L, Li L. 2012b. Pressure-assisted capillary electrophoresis coupling with matrix-assisted laser desorption/ionization-mass spectrometric imaging for quantitative analysis of complex peptide mixtures. Anal Chem 84:7684–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Lee CY, Croushore CA, Sweedler JV. 2012. Label-free quantitation of peptide release from neurons in a microfluidic device with mass spectrometry imaging. Lab Chip 12:2037–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Frost DC, Li L. 2019. High-Resolution Enabled 5-plex Mass Defect-Based N,N-Dimethyl Leucine Tags for Quantitative Proteomics. Analytical Chemistry 91:7991–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Frost DC, Yu Q, Li M, Gu TJ, Li L. 2020. Mass Defect-Based DiLeu Tagging for Multiplexed Data-Independent Acquisition. Anal Chem 92:11119–11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Yu Q, Ma F, Frost DC, Lu L, Chen Z, Zetterberg H, Carlsson C, Okonkwo O, Li L. 2019. HOTMAQ: A Multiplexed Absolute Quantification Method for Targeted Proteomics. Analytical Chemistry 91:2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Liotta LA, Petricoin EF. 2012. The spectra count label-free quantitation in cancer proteomics. Cancer Genomics Proteomics 9:135–142. [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Mabrouk OS, Kennedy RT. 2013. Rapid preconcentration for liquid chromatography-mass spectrometry assay of trace level neuropeptides. J Am Soc Mass Spectrom 24:1700–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wong JM, Mabrouk OS, Kennedy RT. 2015. Reducing adsorption to improve recovery and in vivo detection of neuropeptides by microdialysis with LC-MS. Anal Chem 87:9802–9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilch LW, Kaleta DT, Kohtani M, Krishnan R, Jarrold MF. 2007. Folding and unfolding of helix-turn-helix motifs in the gas phase. J Am Soc Mass Spectrom 18:1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman TA, Rubakhin SS, Romanova EV, Tucker KR, Sweedler JV. 2009. MALDI mass spectrometric imaging using the stretched sample method to reveal neuropeptide distributions in aplysia nervous tissue. Anal Chem 81:9402–9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman TA, Rubakhin SS, Sweedler JV. 2011. MALDI mass spectrometry imaging of neuronal cell cultures. J Am Soc Mass Spectrom 22:828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. 2000. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem 72:563–573. [DOI] [PubMed] [Google Scholar]