Figure 4. Intrinsic defects in aged ILC2s predispose to cold intolerance.
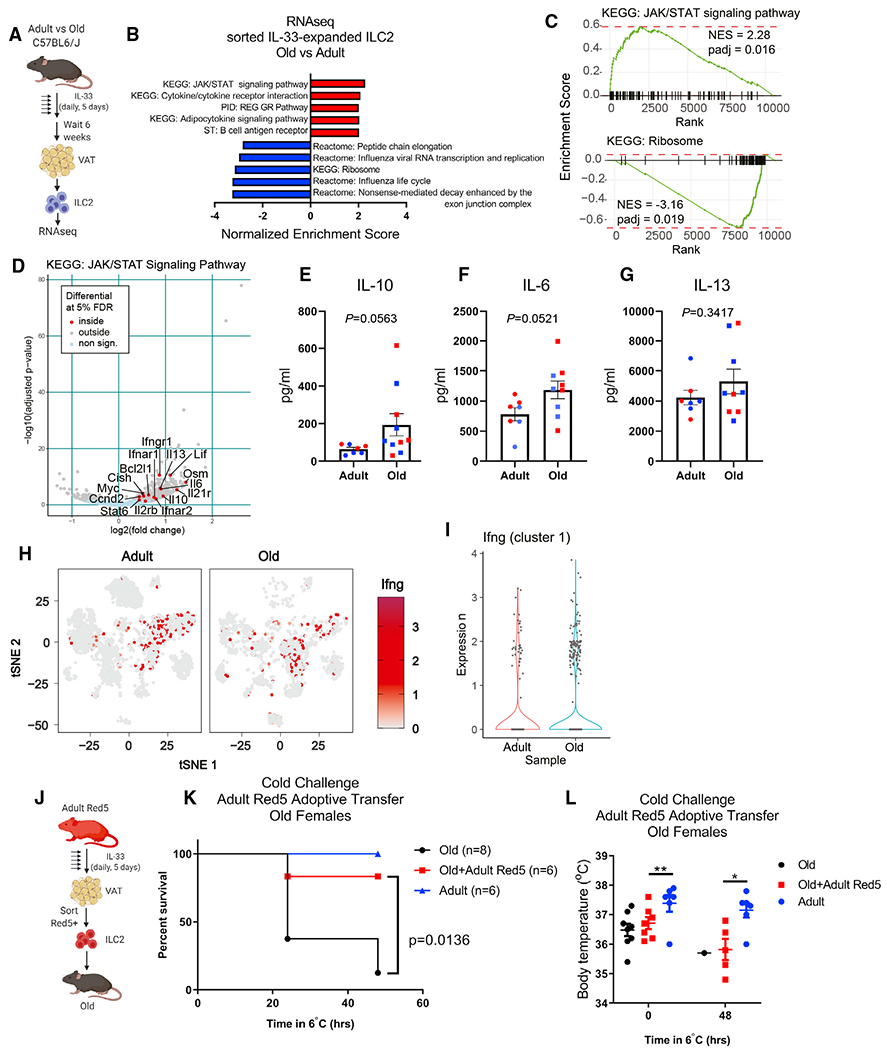
(A) ILC2 bulk RNA-seq experimental design. IL-33 was given by i.p. injection daily for 5 consecutive days and mice were rested for 6 additional weeks prior to tissue collection and ILC2 isolation.
(B) Gene set enrichment analysis of IL-33-expanded ILC2s from adult and old mice. n = 3 adult and n = 3 old samples, each containing sorted ILC2s pooled from n = 2 biological replicate male mice were used for bulk RNA-seq analyses.
(C) GSEA curves of significantly regulated pathways identified in (B).
(D) Volcano plot highlighting significantly regulated genes from the KEGG JAK/STAT signaling pathway gene list.
(E–G) (E) IL-10, (F) IL-6, and (G) IL-13 cytokine secretion was measured from sorted adult and old adipose tissue ILC2s after IL-33-mediated expansion in vivo.
Each symbol represents an individual mouse, and both age groups are pooled from male (blue) and female (red) mice. All mice were treated with IL-33 simultaneously, ILC2s were sorted from n = 1–2 mice/group and stimulated across 4 separate days, and all supernatants were analyzed together. Statistical differences were calculated by t test. Error bars are SEM.
(H) Ifng expression is depicted in red and overlaid across all tissue-resident CD45+ cell subsets in gray.
(I) Violin plots of Ifng expression within cluster 1 αβ T cells.
(J) Adoptive transfer experimental design. Red5 mice were treated with IL-33 by i.p. injection daily for 5 consecutive days.
(K) Survival of old female mice that survived 48 h cold challenge ± receiving adult female Red5+ cells. Date are pooled from two independent experiments. Statistical differences were analyzed by log-rank test.
(L) Body temperatures of old female mice that survived 48 h cold challenge ± receiving adult female Red5+ cells. Data are pooled from 2 independent experiments. Statistical differences in body temperature were calculated by two-way ANOVA. Error bars are SEM.
