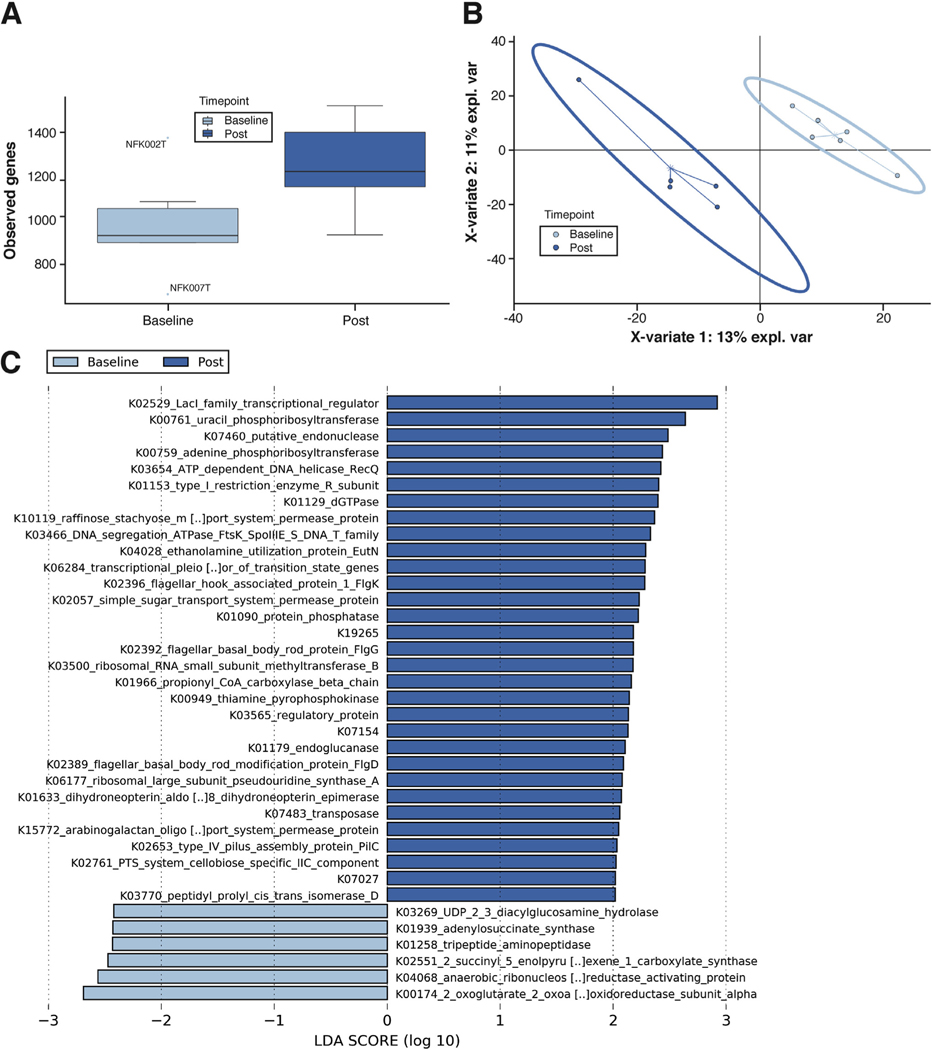
Figure 1.
Metatranscriptomics demonstrates that aerobic exercise training improves gut dysbiosis. (A) α-Diversity values when considering the observed features metric. The postexercise samples had a noticeably higher number of observed genes. Values were calculated using Qiime2 and represent the average of each sample’s 20 iterations at a depth of 12,000 sequences. (B) β-Diversity assessed by multidimensional plots using partial least-squares discriminant analysis, as implemented in the mixOmics package in R software, shows separation between groups in both 2-dimensional space (shown) and 3-dimensional space (not shown). (C) LEfSe enrichment plots display significantly enriched (P < .05; LDA > 2) functional KEGG genes within the groups. The logs of LDA scores are displayed on the x-axis and quantify the strength of enrichment within each respective categorical group. LEfSe assesses significance using Kruskal-Wallis tests.