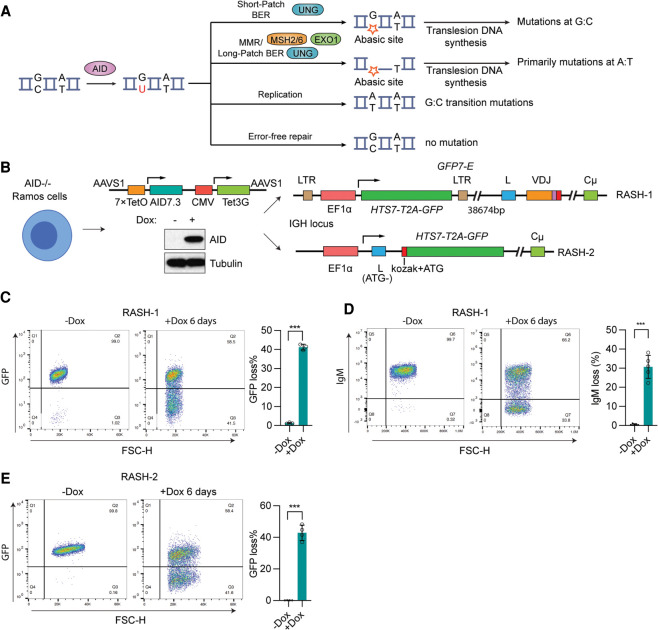
Figure 1.
Establishing a rapid assay for SHM. (A) Schematic of SHM reaction. During SHM, AID deaminates C to generate uracil, which can act as a template for replication, leading to a C:G-to-T:A transition mutation. Alternatively, noncanonical (error-prone) base excision repair (BER) initiated by UNG can create an abasic site that, if copied by a translesion DNA polymerase, yields mutations at G:C. Finally, U or U:G mismatches can trigger the generation of ssDNA gaps through the action of UNG and mismatch repair (MMR) factors MSH2, MSH6, and EXO1, which primarily yield mutations at A:T upon translesion polymerase synthesis. (B) Scheme for creating RASH-1 and RASH-2. Creation of RASH involved several steps of CRISPR/Cas9 gene targeting in Ramos. AICDA (encoding AID) was disrupted, and a construct was inserted into the AAVS1 safe harbor locus that expresses AID7.3 (AID hyperactive mutant with threefold increased catalytic activity) upon doxycycline (dox)-induced binding of Tet3G to a promoter containing seven copies of its binding site, TetO (7xTetO). Western blot showing AID protein levels in AID7.3in cells without or with 200 ng/mL dox for 24 h. The GFP7-E reporter with EF1α promoter driving transcription of HTS7 (enriched with AID hotspots)-T2A-GFP was integrated into IGH either 38,674 bp upstream of the V leader sequence (RASH-1) or by replacing IGH-VDJ coding sequences so as to retain the leader exon (which now lacks its ATG start codon; RASH-2). Kozak sequence was added to HTS7-T2A-GFP in RASH-2. (C,D) Representative flow cytometry plots of and bar graphs quantifying GFP loss (C) or IgM loss (D) in RASH-1 cells treated with dox for 6 d. (E) Representative flow cytometry plots of and bar graph quantifying GFP loss in RASH-2 treated with dox for 6 d. Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using two-tailed Student's t-test. (***) P value < 0.001.