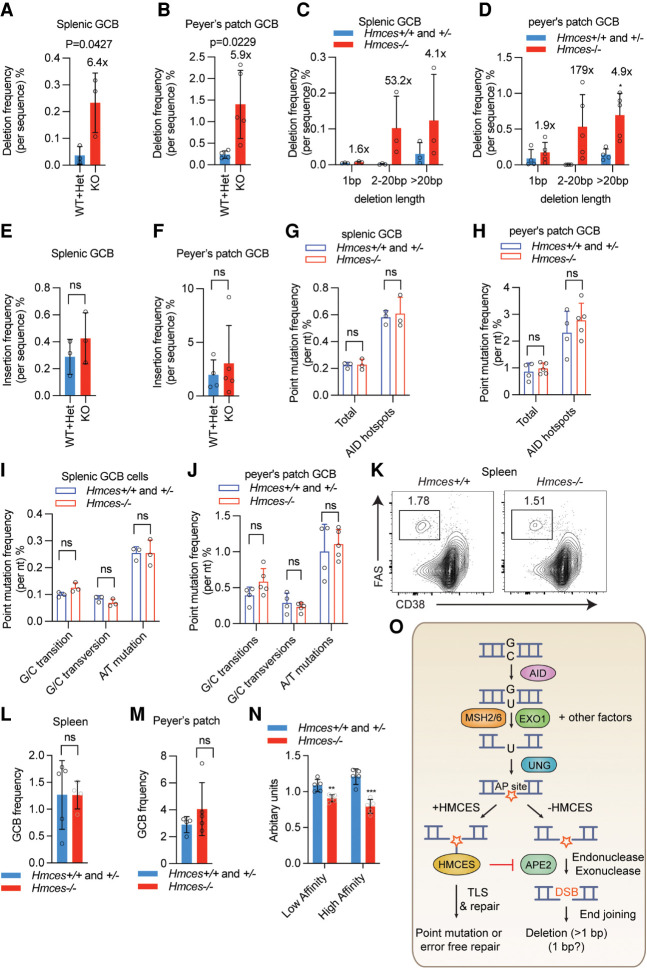
Figure 7.
HMCES deficiency leads to increased deletions in mouse germinal center B cells. (A,B) Deletion frequencies in the region downstream from the Igh J4 gene segment (JH4 intron) in germinal center B (GCB) cells from either the spleen (A) or Peyer's patch (B) of Hmces−/− and control Hmces+/− and Hmces+/+ mice. (C,D) Distribution of deletion lengths in the JH4 intron in splenic (C) and Peyer's patch GCB (D) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (E,F) Insertion frequencies in the JH4 intron in splenic (E) and Peyer's patch GCB (F) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (G,H) Point mutation frequencies in the JH4 intron in splenic (G) and Peyer's patch GCB (H) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (I,J) Mutation frequencies for G/C transitions, G/C transversions, and A/T mutations in the JH4 intron in splenic (I) or Peyer's patch GCB (J) cells from Hmces−/− and control Hmces+/+ and Hmces+/− mice. (K) Flow cytometry plots of GCB cells gated as the FAS+ CD38− population among total B cells in spleens of Hmces+/+ and Hmces−/− mice 14 d after immunization with NP-CGG. (L,M) Bar graphs showing the frequencies of GCB cells in Hmces+/+ or Hmces+/− and Hmces−/− mice in spleens (L) or Peyer's patches (M). (N) Bar graphs quantifying the levels of low- and high-affinity IgM antibodies measured by ELISA in the sera of Hmces−/− and control Hmces+/+ and Hmces+/− mice. NP-BSA with a conjugation ratio of 27 and 2 was used to detect low-and high-affinity antibodies, respectively. (O) Working model for the mechanism by which HMCES suppresses deletions during SHM. After AID acts, MSH2/6, EXO1, and likely other factors can resect one strand to expose a region of ssDNA containing a U residue, on which UNG can act to create an abasic site (AP site). Cleavage of this vulnerable intermediate by APE2 would result in a DNA double-strand break (DSB), which would be prone to end resection by exonucleases (including APE2 itself) and to generating a deletion. HMCES protects against this outcome by forming a covalent cross-link with the abasic site, thereby blocking cleavage by APE2. The events that occur subsequent to HMCES cross-linking are not known but could involve translesion synthesis (TLS) by an error-prone polymerase. The pathway depicted here is just one of the pathways depicted in Figure 1A (the second from the top). Throughout the figure, data are presented with the bars representing mean and the error bars as ±SD. Statistical significance was calculated using two-tailed Student's t-test. (***) |P-value < 0.001, (**) P-value < 0.01, (ns) not significant.