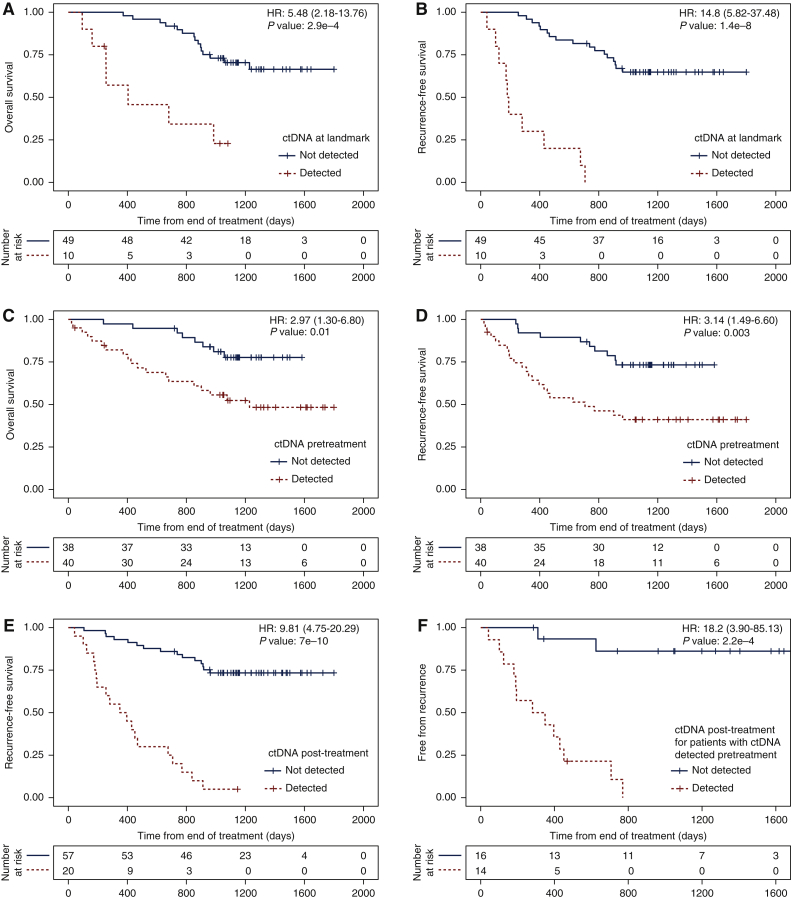
Figure 4.
Survival analysis based on ctDNA detection.
Kaplan–Meier analysis showing the fraction of patients without events as a function of time. Patients right-censored due to loss of information are shown as vertical tick-marks. Patient subgroups are defined based on ctDNA detection using the RaDaR™ assay at different timepoints (see individual panels). Patients with ctDNA detected are shown in red dotted lines, and those with ctDNA not detected at the respective timepoints are shown in black solid lines. The numbers of patients remaining at risk are shown below each graph. (A) Overall survival (OS) and (B) recurrence-free survival (RFS, counting as events either recurrence of the first primary tumour or death if not preceded by a second primary tumour), for patients split by ctDNA detection at the landmark timepoint, which is the first plasma sample available in the window ≥2 weeks and ≤4 months after the end of treatment (data available for 59 patients in total). Hazard ratios: 5.48 [95% confidence interval (CI): 2.18-13.76] and 14.8 (95% CI: 5.82-37.48) for OS and RFS, respectively (P values: 2.9e-4 and 1.4e-8). (C) OS and (D) RFS for patients split by ctDNA detection before treatment (data available for 78 patients in total). Hazard ratios: 2.97 (95% CI: 1.30-6.80) and 3.14 (95% CI: 1.49-6.60) for OS and RFS, respectively (P values: 0.01 and 0.003). (E) RFS for patients split by ctDNA detection at any timepoint ≥2 weeks after the end of treatment (data available for 77 patients in total). Hazard ratio: 9.81 (95% CI: 4.75-20.29, P value: 7e-10). (F) Fraction of patients who remain free from recurrence split by ctDNA detection at any timepoint ≥2 weeks after the end of treatment, for patients for whom ctDNA was detected before treatment (data available for 30 patients in total). Hazard ratio: 18.2 (95% CI: 3.9-85.13, P value: 2.2e-4).