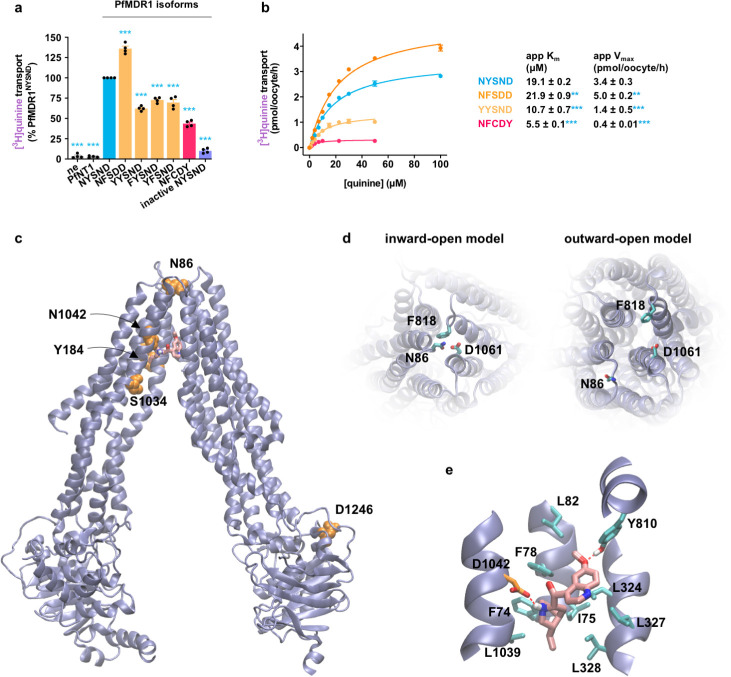
Fig 4. Quinine transport via PfMDR1.
(a) Field isoforms of PfMDR1 transported [3H]quinine, with PfMDR1NFSDD exhibiting the highest level of quinine transport activity. (b) There were significant differences in the apparent kinetic parameters for quinine transport via field isoforms of PfMDR1. The concentration dependence of PfMDR1-mediated quinine transport was calculated by subtracting the leakage from ne from that of oocytes expressing a PfMDR1 isoform at each drug concentration. The data are the mean of n = 4 independent experiments (each yielding similar results and overlaid as individual data points in panel a), and the error is the SEM. The asterisks denote a significant difference from PfMDR1NYSND; **P < 0.01 and ***P < 0.001 (1-way ANOVA). The data underlying panels a and b of this figure is supplied in S3 Data. (c) Inward-open homology model of PfMDR1 based on the C. elegans P-gp crystal structure (PDB 4F4C) showing the location of the mutations found in the 5 field PfMDR1 isoforms characterized in this study (orange). A putative binding pose of quinine in the central cavity is shown (pink). (d) A comparison of the inward-open model and outward-open models (based on the crystal structure of human P-gp; PDB 6C0V) as viewed from the side of the protein facing into the DV lumen. The N86Y mutation is part of a cluster of 3 residues forming the extracellular gate of the transporter (the participating residues are shown). These 3 residues are in close proximity in the inward-open state but move apart in the outward-open conformation. (e) Putative quinine binding site in PfMDR1NFSDD. Amino acids interacting with quinine are indicated (apart from L71, I1071, and F1072 that are removed to clearly view the binding pose). Atoms are shaded as follows: carbon in quinine, pink; carbon in the 1042D residue, orange; nitrogen, blue; oxygen, red; hydrogen, white. ne, nonexpressing oocytes; PfMDR1, Plasmodium falciparum multidrug resistance protein 1; P-gp, P-glycoprotein.