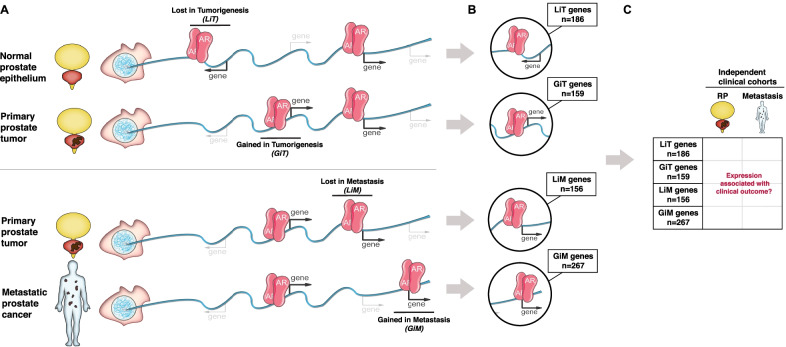
Fig. 1.
Schematic of the study methodology. A Schematic of AR biding sites unique to specific clinical states. Depicted are AR sites specific to healthy prostate epithelium and lost in primary prostate tumors (LiT)—i.e., AR is present in the first DNA strand representing the normal prostate genome and absent in the second strand representing the localized prostate tumor genome; AR sites absent in primary prostate epithelium and gained in primary prostate tumors (GiT); AR sites present in primary prostate tumors and lost in prostate cancer metastases (LiM); and AR sites absent in primary prostate tumors and gained in prostate cancer metastases (GiM). B Gene sets were selected based on their differential expression across tumor types and their proximity (≤ 50 kb) to tissue-specific AR sites. The gray arrows from each DNA strand direct to a highlighted region from that strand exemplifying a context-specific AR site (LiT, GiT, LiM, or GiM) and its associated gene. The number of genes identified genome-wide from each category are indicated in the text box linked to the representative genomic regions. C Clinical outcome based on expression of the genes within each individual cohort (LiT, GiT, LiM, or GiM along the y-axis) was examined in two independent cohorts: a localized disease cohort (RP, radical prostatectomy) and a metastatic cohort (metastasis) as depicted across the x-axis. In total, eight independent analyses were performed