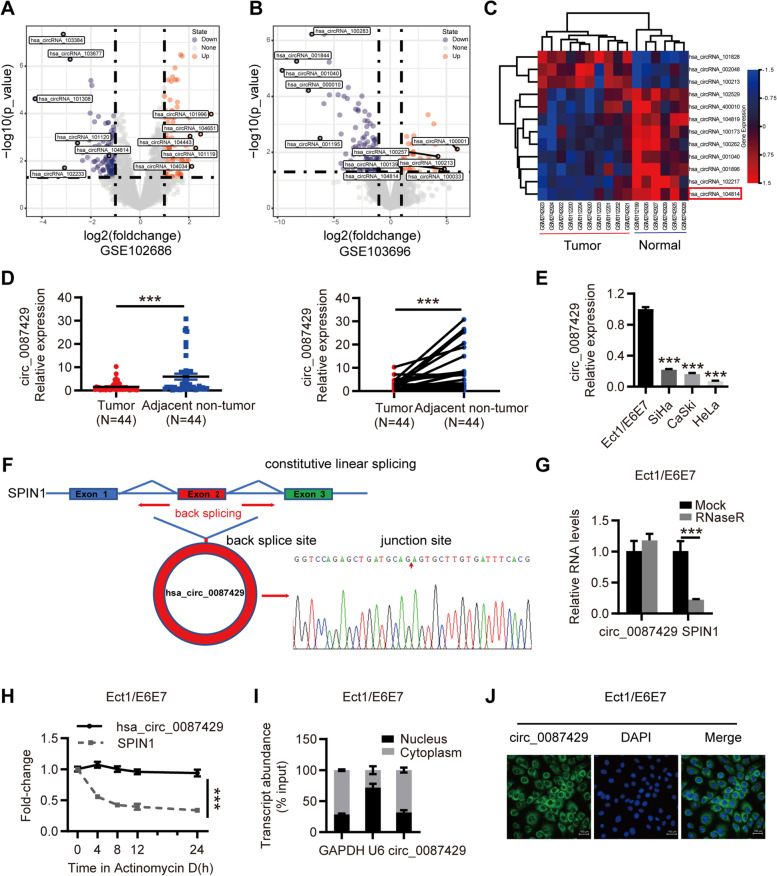
Fig. 1.
The expression and characteristics of circ_0087429 in cervical cancer. a. Differentially expressed circRNAs between cervical cancer and adjacent tissue samples in the GSE102686 dataset. b. Differentially expressed circRNAs between cervical cancer and normal cervical epithelial cell lines in the GSE103696 dataset. c. Heatmap of circRNAs with significant differential expression in two datasets (GSE102686 and GSE103696). circ_0087429 (hsa_circRNA_104814) is marked with a red box. d. Differential expression of circ_0087429 in 44 pairs of cervical cancer and adjacent tissue samples. e. Differential expression of circ_0087429 between cervical cancer and normal cervical epithelial cell lines. f. The Sanger sequencing results showed that circ_0087429 was formed by head-to-tail splicing of the second exon of the parental gene SPIN1. g. The expression of circ_0087429 and SPIN1 after RNaseR treatment was detected by qRT-PCR. h. The expression changes in circ_0087429 and SPIN1 at different time points after adding actinomycin D were determined by qRT-PCR. i. A subcellular fractionation assay was used to detect the relative expression of circ_0087429 in the cytoplasm and nucleus. U6 was used as the internal reference for the nuclear fraction, and GAPDH was used as that for the cytoplasmic fraction. j. FISH staining confirmed the expression of circ_0087429 in the cytoplasm. Scale bar, 100 μm. ***p < 0.001