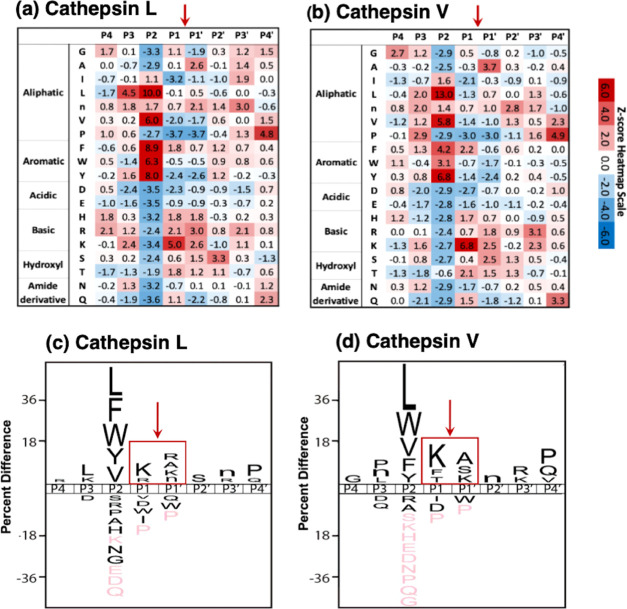
Figure 3.
Cathepsin L and cathepsin V preferences for P4–P4′ residues of peptide cleavage sites in MSP-MS analyses. (a, b) Heat maps of amino acids preferred at cleavages sites. The peptide library cleavage data for cathepsin L (panel a) and cathepsin V (panel b) shows the frequencies of amino acid residues at each of the P4 to the P4′ positions of cleaved peptides, shown as the heat maps of Z-scores (explained in the Methods section) that compare protease cleavages with that of the reference peptide library. (c, d) IceLogo of cathepsin L and cathepsin V for preferred cleavages at P4–P4′ residues. Cathepsin L (panel c) and cathepsin V (panel d) cleavage data is illustrated by iceLogo. IceLogo shows the relative frequency of the preferred residues at the P1–↓P1′ cleavage site and at the P4–P4′ residues. Black letters above the line of P4–P4′ positions indicate preferred amino acid residues of the protease with p < 0.05, compared to the reference (negative data) of all possible residues at each position. Pink letters indicate residues that were never found at the indicated cleavage position.