Fig. 2. METTL16 enhances translation efficiency of target mRNAs.
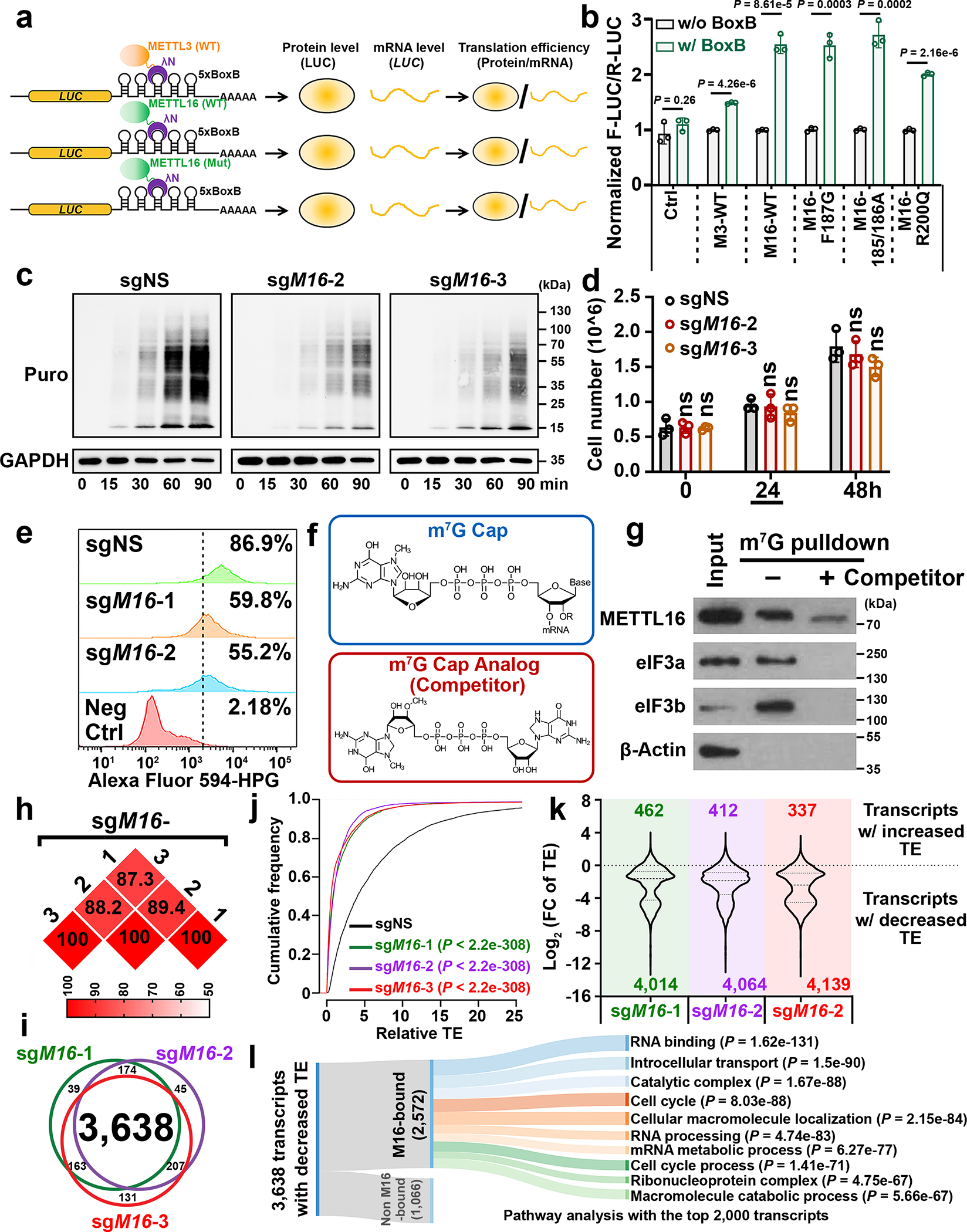
a, Experimental schedule of RNA tethering experiments to determine translation efficiency. b, Translation efficiency as determined by the tethering assays. F-Luc, Firefly luciferase; R-Luc, Renilla luciferase, internal control. Data are mean ± s.d. (n = 3 independent experiments). Statistics: unpaired, two-tailed t-test. c, Representative Western blotting images of SUnSET assays to quantify the amount of nascent (puromycin-labelled) peptides in HEK293T cells with METTL16 knockout (KO). To make the data comparable, all the samples were treated equally. Data shown represent 3 independent experiments. d, The cell number in sgNS, sgM16-2, and sgM16-3 groups. The samples were collected at 24 hours for the SUnSET assay shown in Fig. 2c. Data are mean ± s.d. (n = 3 independent experiments). Statistics: unpaired, two-tailed t-test. ns, not significant (P ≥ 0.05). e, Nascent protein synthesis upon METTL16 KO as determined by the pulse labeling assay via incorporation of L-homopropargylglycine (HPG). f, Structures of m7G cap and its analog. Data shown represent 2 independent experiments. g, Western blotting of m7G pulldown assays with indicated antibodies. eIF3a and eIF3b, positive controls. β-Actin, negative control. Data shown represent 2 independent experiments. h, The reproducibility of Ribo-seq with three diverse gRNAs against METTL16. Here, we presented the percentages of overlapping transcripts with decreased translation efficiency (TE) caused by the three gRNAs mediated METTL16 KO in HEK293T. i, Venn diagram showing high overlap of the transcripts with inhibited TE across the three gRNA groups. j, CDF plot depicting the transcriptome-wide inhibition of translation efficiency (TE) upon METTL16 KO. Statistics: two-sided Wilcoxon and Mann–Whitney test. k, Violin plots showing the Log2(FC of TE) upon METTL16 KO. FC, fold change. l, Sankey diagram presenting the subsequent pathway analysis of the transcripts whose translation initiation is facilitated by METTL16. Among the 3,638 transcripts with suppressed TE upon METTL16 KO, 2,572 are directly bound by METTL16 protein. Among the 2,572 targets, we selected the top 2,000 candidates for pathway analysis. Here, we showed the top 10 enriched pathways.
