Fig. 5. METTL16 facilitates the formation of TIC by interactions with both eIF3a/b and rRNAs.
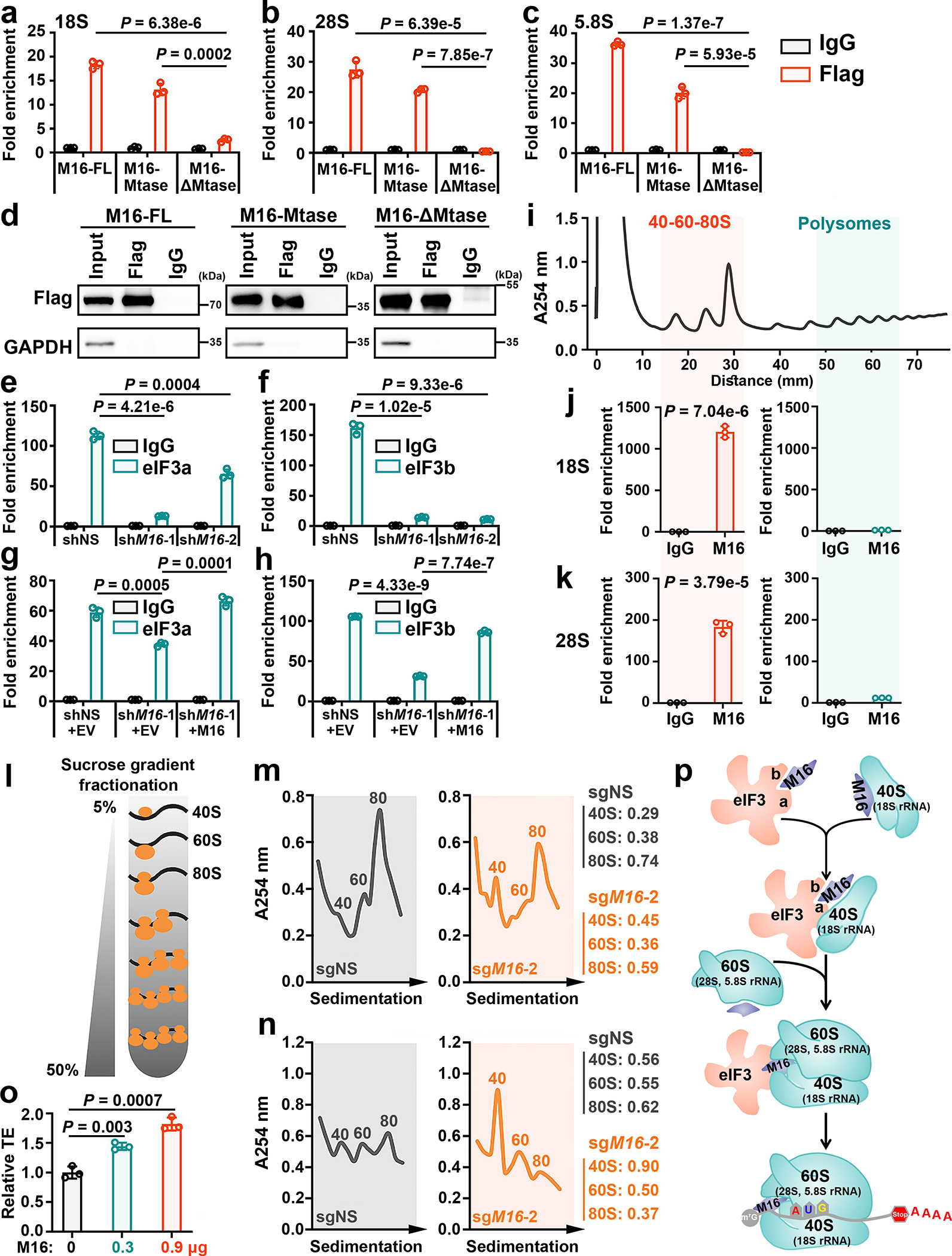
a, The binding of full length (FL), Mtase domain, and ΔMtase domain of METTL16 on 18S (a), 28S (b), and 5.8S (c) rRNAs in the cytoplasm fraction as determined by CLIP-qPCR. d, The comparable pulldown efficiency among METTL16-FL, Mtase, and ΔMtase groups. Data shown represent 3 independent experiments. e, f, CLIP-qPCR analysis showing the decreased association between 18S rRNA and eIF3a (e) or eIF3b (f) upon METTL16 KD. g, h, Rescue assay showing the rescued binding of eIF3a (g) or eIF3b (h) with 18S rRNA upon manipulating the expression of METTL16. i, Isolation of translation initiation fraction (40–60-80S) and polysome fraction for RIP according to polysome profiling. Data shown represent 3 independent experiments. j, k, The interactions between METTL16 and 18S rRNA (j) or 28S rRNA (k) in the translation initiation fraction (left) and polysome fraction (right). HEK293T cells were used in all the above studies. l, Schematic of sucrose gradient fractionation. m, n, Sucrose density gradient profiles showing the abundance of 40S, 60S, and 80S ribosomes from HEK293T (m) and HepG2 (n) cells upon METTL16 KO. Data shown represent 3 independent experiments. o, Effect of ectopic METTL16 protein on translation efficiency of luciferase mRNA as determined by in vitro translation. p, Schematic model of how METTL16 enhances translation initiation. The direct interactions of METTL16 with eIF3a/b and rRNAs drive the eIF3/40S ribosomal subunit (with 18S rRNA as the core) interaction and the subsequent assembly of 40S ribosomal subunit/60S ribosomal subunit (with 5.8S and 28S rRNAs as the core) into the 80S TIC, leading to stimulated translation initiation. For a-c, e-h, j, k, and o, data are represented as mean ± s.d. (n = 3 independent experiments). Statistics: unpaired, two-tailed t-test. For j and k, the P values indicate the binding of METTL16 with 18S rRNA (j) and 28S rRNA (k) in 40–60-80S vs Polysomes.
