Extended Data Fig. 3. METTL16 specifically deposits m6A RNA methylation in a set of mRNA targets and its biological role in cell proliferation/growth and translation promotion can’t be substituted by METTL3 and METTL14.
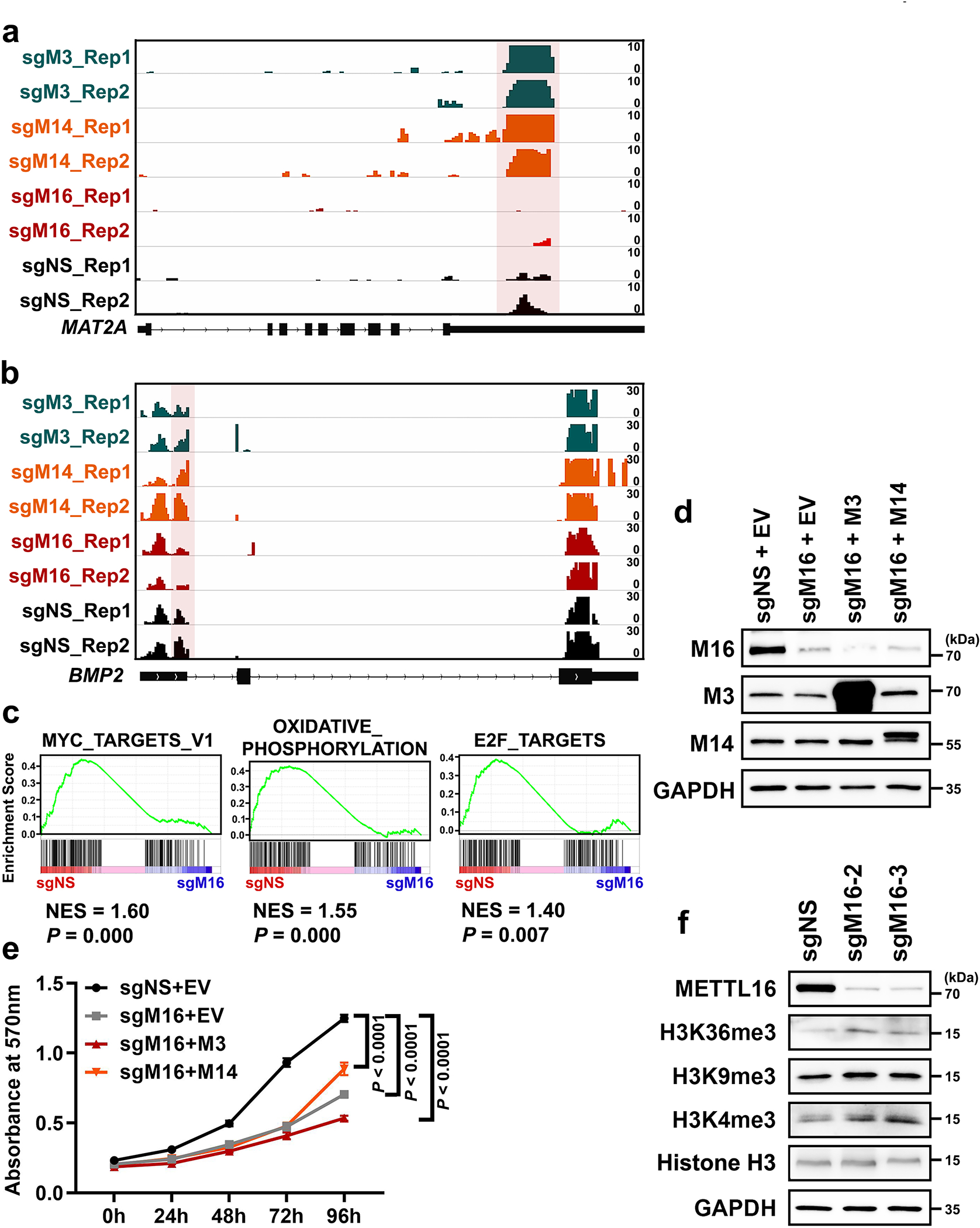
a, b, Integrative Genomics Viewer (IGV) tracks displaying the m6A read distribution and changes (based on the m6A MeRIP-seq data) in MAT2A (a) and BMP2 (b) mRNA upon the KO of METTL3, METTL14, or METTL16 in HEK293T cells. The m6A decorations in MAT2A and BMP2 mRNA are METTL16-dependent, and METTL3/14-independent. c, The top 3 signaling pathways suppressed by METTL16 KO (sgMETTL16-3) as determined by GSEA. d, Overexpression efficacy of METTL3 and METTL14 in HEK293T cells with endogenous METTL16 KO (sgMETTL16-2). Images are representative of 2 biologically independent experiments with similar results. e, Effects of METTL3 and METTL14 overexpression on cell growth in HEK293T cells with endogenous METTL16 KO (sgMETTL16-2). Data are plotted as mean ± s.d. (n=3 independent experiments). Statistics: two-way ANOVA. f, Assessment of the global level changes of histone methylations, including H3K36me3, H3K9me3, and H3K4me3, in HEK293T cells upon endogenous METTL16 KO by Western blotting. Images are representative of 2 biologically independent experiments with similar results.
