Fig. 4 |. Single-cell transcriptomic analysis reveals the presence of diverse cellular populations in fusion organoids with a trend towards accelerated maturation and alterations in interneuron formation in MECP2 mutant samples.
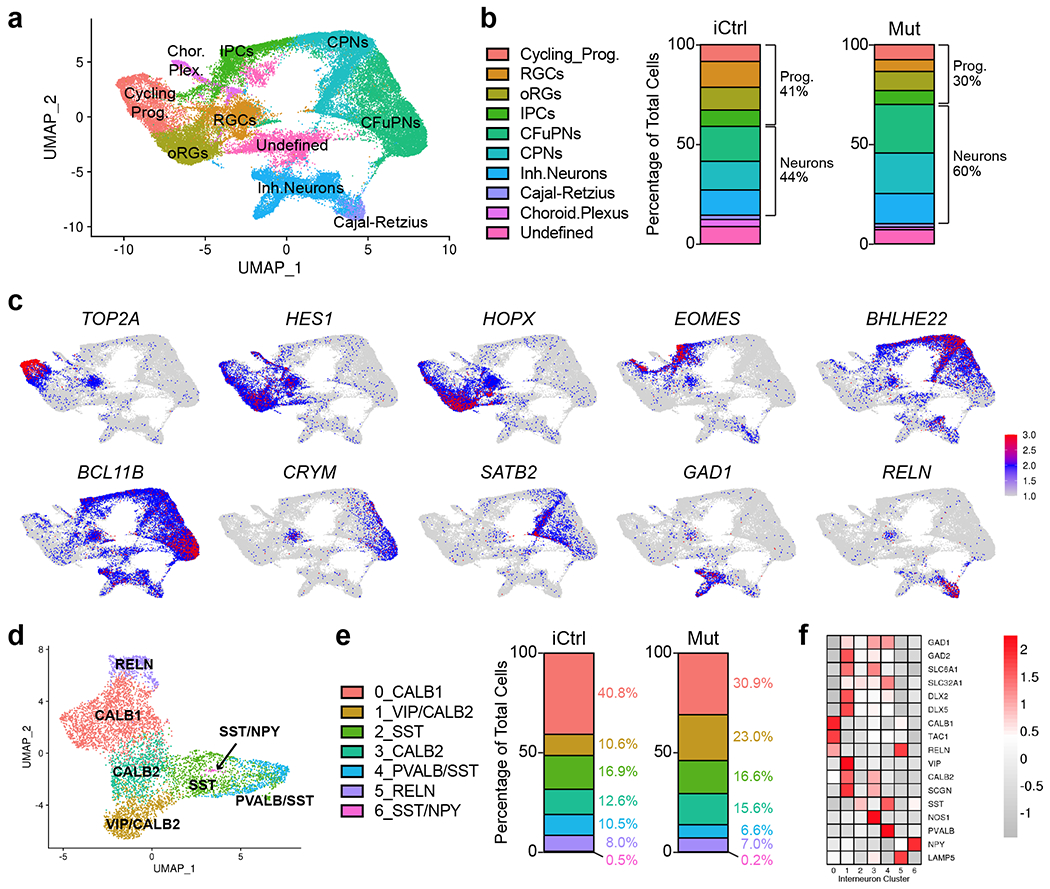
a, Uniform Manifold Approximation and Projection (UMAP) of combined iCtrl and Mut Cx and GE organoids. The plot includes cells from 3 D56 Cx and 3 GE organoids collected before fusion, 3 D70 Cx+GE fusion organoids, and 3 D100 Cx+GE fusion organoids. The total number of cells sequenced were as follows: D56 iCtrl, 9306; D56 Mut, 9186; D70 iCtrl, 10931; D70 Mut, 6260; D100 iCtrl, 7561; and D100 Mut, 6698 cells. b, Plots display the mean percentage of cells in the fusion organoids representing each of the clusters in a. Separation of the data by iCtrl and Mut status shows a trend of reduced progenitors and more differentiated neurons in Mut organoids compared to iCtrl samples. c, UMAPs of key genes associated with each of the major clusters identified in a. d, Re-clustered UMAP of the interneuron subset from a with interneuron subtype markers identifying each re-clustered subset. e, Percentage of cells for each of the clusters in d segregated by iCtrl and Mut reveals increased numbers of interneurons expressing PVALB/SST and CALB1 in iCtrl organoids and cells expressing VIP and CALB2 in Mut samples. f, Heat map with the relative expression of canonical interneuron-related genes within the re-clustered groups.
