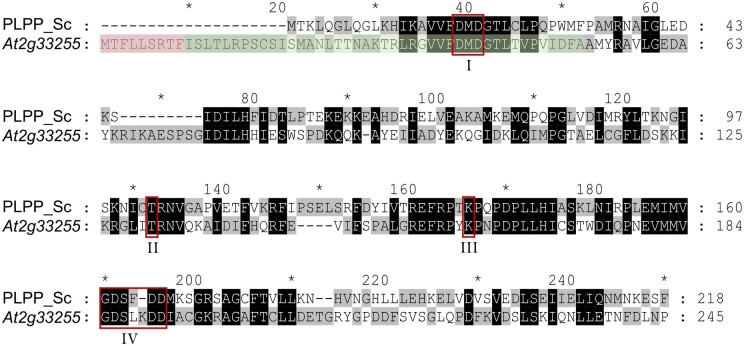
Figure 5.
ClustalW alignment of the amino acid sequences of the PLPP from S. cerevisiae (PLPP_Sc; YOR131C) and the At2g33255 gene product. Residues identical among the two sequences or with similar properties are indicated by black shading. Residues not identical are highlighted by different shading (gray/white). Red boxes mark conserved HADSF motifs I–IV (I: DxD; II, T/S; III: K/R; IV: E/DD, GDxxxD, or GDxxxxD). The putative mitochondrial targeting sequence (according to TargetP) is shaded in red. The predicted chloroplast targeting sequence (according to ChloroP) comprises also the green shaded residues. Dashes represent introduced gaps for alignment improvement. Numbers at the right indicate amino acid positions. The amino acid sequence alignment also demonstrates that the At2g33255 encoded protein is N-terminally extended by 20 residues when compared with the cytosolically located PLPP YOR131C from S. cerevisiae. Such extensions often act as targeting sequences signifying the import of proteins into mitochondria or chloroplasts. Notably, the At2g33255 encoded protein does not share substantial sequence similarity (25%) to a recently identified chloroplastic PLP phosphatase from N. tabacum, named NtPLPP1 (ShuoHao et al., 2019; Supplemental Figure S5).