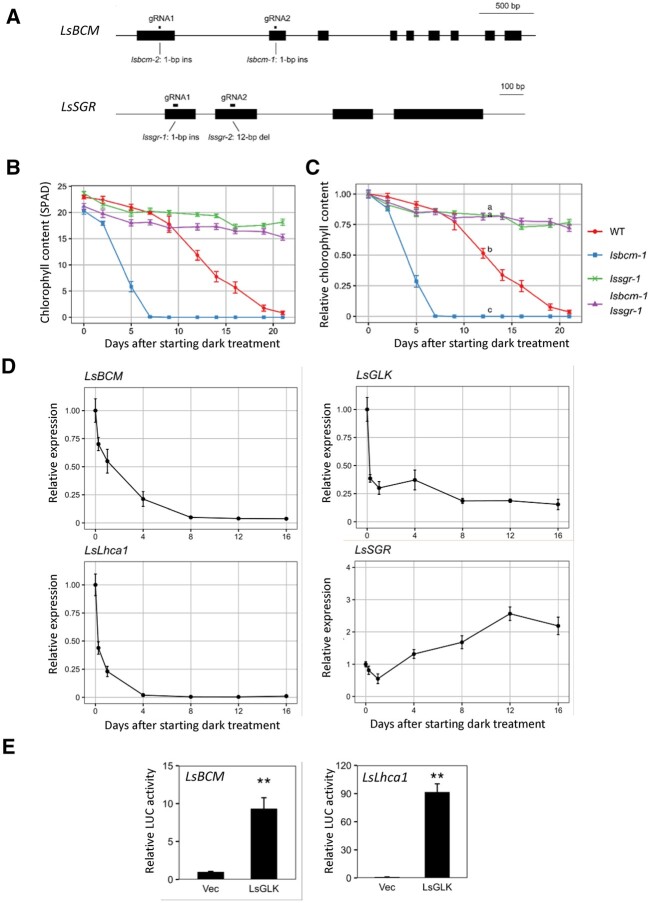
Figure 4.
Functional analysis of the BCM ortholog in lettuce. A, Structures of lsbcm-1, lsbcm-2, lssgr-1, and lssgr-2 mutants generated using CRISPR-Cas9-based genome editing. lsbcm-1 and lsbcm-2 were obtained using gRNA1 in the first exon and gRNA2 in the second exon. lssgr-1 and lssgr-2 were obtained using gRNA1 in the first exon and gRNA2 in the second exon. B and C, Changes in Chl content with time during dark incubation. Third leaves of three-week-old plants incubated at 22°C in the dark were used. B and C indicate SPAD values and relative values to presenescent leaves, respectively. D, Changes in gene expression of LsBCM, LsGLK, LsLhca1, and LsSGR with time. Lettuce leaves were incubated in the dark as described in panel B. E, Transactivation of LsBCM and LsLhca1 expression by LsGLK1. Constructs carrying luciferase genes driven by promoters of LsBCM1 and LsLhca1 were transiently introduced into lettuce mesophyll protoplasts together with LsGLK1 constructs. “Vec” represents a control into which the luciferase reporter construct and an empty effector vector were introduced. n = 3. **P < 0.01 (Student’s t test).