Figure 3.
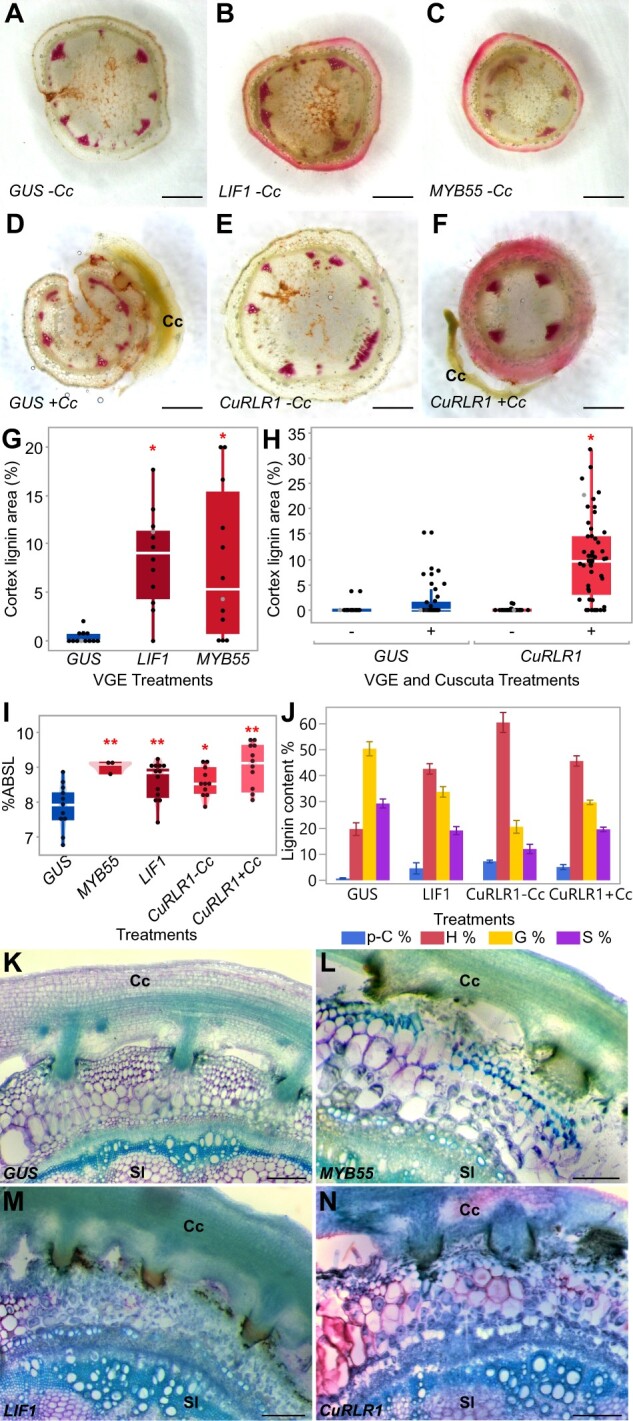
VGE in tomato H1706. A–F, ∼300-μm stem sections near injection sites, Ph–HCl stains lignin red. Scale bar, 1 mm. VGE of GUS (A), LIF1 (B), SlMYB55 (C), and CuRLR1 (E) in stem without C. campestris. VGE of GUS (D) and CuRLR1 (F) with C. campestris. G, Cortex lignin area percentage in VGE of LIF1 and SlMYB55 (n = 12 each) and H, CuRLR1 with and without C. campestris (GUS-Cc, n = 18; GUS+Cc, n = 28; CuRLR1−Cc, n = 47; CuRLR1+Cc, n = 53). G and H, Data were collected at 7-d post injection (DPI) and 14 DPA. Data are assessed using Dunnett’s test with GUS−Cc as negative control. *P < 0.01. The data points labeled with grey color indicate the sample that we show in the section picture (A–F). I, Acetyl bromide assay for lignin in VGE stems. Acetyl bromide soluble lignin (ABSL) indicates percent absorbance of soluble lignin. Samples were collected at 7 DPI and 6 DPA. Data are assessed using Dunnett’s test. *P <0.05, **P <0.01. Biological replicates for GUS, n = 18; SlMYB55, n = 10; LIF1, n = 18, CuRLR1-Cc, n = 18; CuRLR1+Cc, n = 18. Technical replicates for acetyl bromide assay; GUS, n = 11; SlMYB55, n = 3; LIF1, n = 13; CuRLR1−Cc, n = 11; CuRLR1+Cc, n = 11. G–I, The boxplot consists of a box extending from the 25th quantile to the 75th quantile. The centerline in the box indicates the median. The length of the box is the IQR, which is the difference between the 25th quantile and the 75th quantile. The whiskers extend from the ends of the box to the outermost data point that falls within 1.5 times of IQR. Points outside of the whiskers are outliers. J, Pyro-GC assay for monolignols in CuRLR1 VGE samples with and without C. campestris. p-C, p-coumaric acid; H, H types of (p-coumaryl alcohol); G, G types of monolignol (coniferyl alcohol); S, S types of monolignol (sinapyl alcohol). Samples were collected at 7 DPI and 6 DPA. Biological replicates collected from first internodes; GUS, n = 8; LIF1, n = 8, CuRLR1-Cc, n = 18; CuRLR1+Cc, n = 18. Pyro-GC assay technical replicates; GUS, n = 3; LIF1, n = 3; CuRLR1-Cc, n = 5; CuRLR1+Cc, n = 5. K–N, VGE of CuRLR1, LIF1 and SlMYB55 induces cortical lignin making H1706 resistant to C. campestris. Scale bar, 30 µm. Samples were collected at 7 DPI and 6 DPA. Cc indicates C. campestris; Sl indicates S. lycopersicum.
