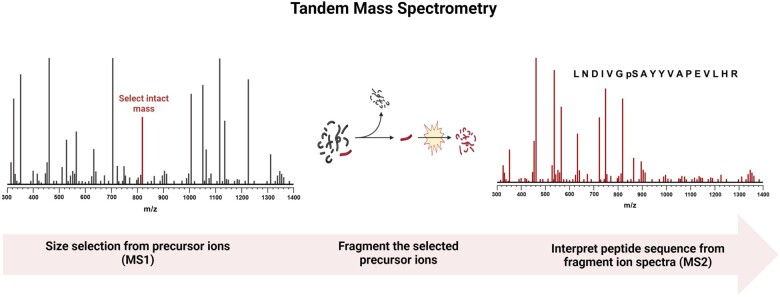
Figure 1.
Principle by which peptides are sequenced via tandem mass spectrometry. First, a mass spectrum (MS1) is obtained to determine which intact masses are present in a sample as it is injected into the mass spectrometer (far left, size select). Next, each identified peptide is independently isolated using a mass filter and fragmented, generally via collision with inert gas molecules, within the instrument (middle, fragment). Finally, the fragment ion spectrum (MS2) is used as a template for matching in silico digested and fragmented proteomes to identify and annotate the sequence including posttranslational modifications (right, interpret sequence).