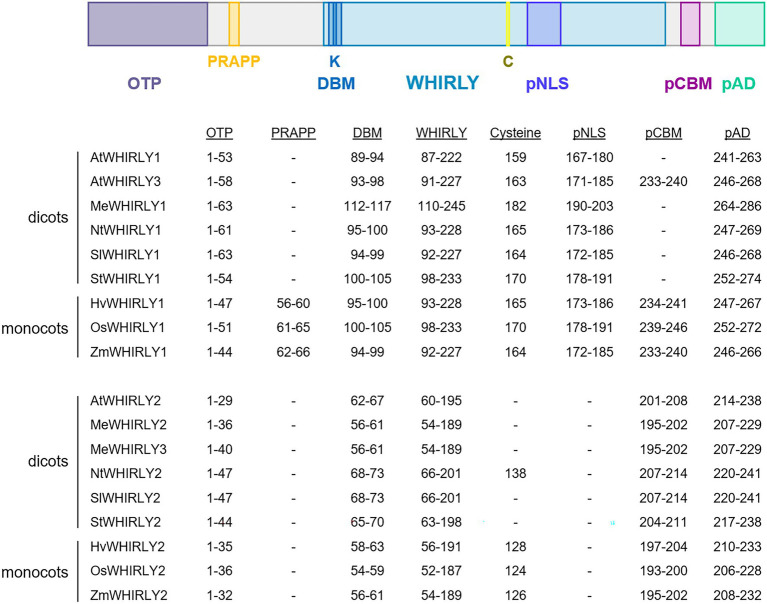
Figure 1.
Schematic comparison of motifs in selected WHIRLY proteins. AtWHIRLY1 (Arabidopsis thaliana, NP_172893), AtWHIRLY2 (A. thaliana, NP_177282.2), AtWHIRLY3 (A. thaliana, NP_178377), HvWHIRLY1 (Hordeum vulgare, BAJ96655), HvWHIRLY2 (H. vulgare, BF627441.2), SlWHIRLY1 (Solanum lycopersicum, AFY24240.1), SlWHIRLY2 (S. lycopersicum, XP_010313085.1), StWHIRLY1 (Solanum tuberosum, NP_001275155.1), StWHIRLY2 (S. tuberosum, NP_001275393.1), NtWHIRLY1 (Nicotiana tabacum, XP_016453689.1), NtWHIRLY2 (N. tabacum, XP_016511175.1), ZmWHIRLY1 (Zea mays, NP_001123589.1), ZmWHY2 (Z. mays, NP_001152589.2), OsWHIRLY1 (Oryza sativa, BAD68418.1), OsWHIRLY2 (O. sativa, NP_001045956.1). The WHIRLY domain is colored in light blue. The N-terminal organelle targeting peptides (OTP) are illustrated in grey. Below the scheme, the positions of selected amino acid residues and motifs in different WHIRLIES are listed: the PRAPP motif in yellow, the DNA-binding motif (DBM) and a putative nuclear localization motif (pNLS) in blue, a putative copper-binding motif in purple and a putative transactivation domain (pAD) in green. The organelle targeting peptides (OTP) were predicted with TargetP-2.0 (https://services.healthtech.dtu.dk/service.php?TargetP-2.0) or UniProt, (https://www.uniprot.org/), the putative nuclear localization signal (pNLS) with NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/) and the putative copper-binding motif (CBM) with Motif Scan (https://myhits.sib.swiss/cgi-bin/motif_scan). The putative autoregulatory domain (pAD) was defined by Desveaux et al. (2005).