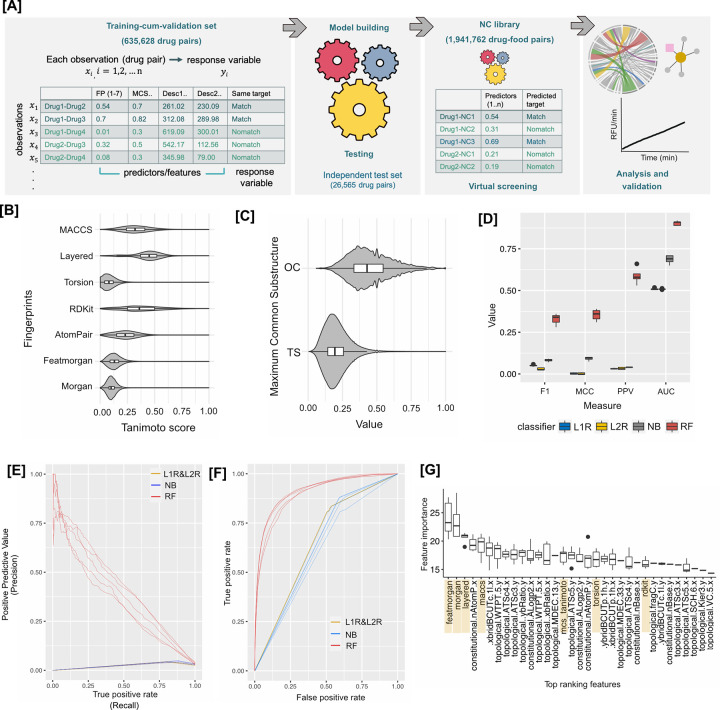
Fig 1.
(A) Overview of workflow deployed. A training-cum-validation set comprising of drug pairs was created using various predictor variables (fingerprints, MCS and physicochemical properties). The model was trained for response variable (Match or Nomatch) and tested on an independent test set for performance evaluation. The natural compound library paired with drugs was virtually screened to obtain hit pairs, followed by analysis and in-vitro validation. (B-C)—Similarity metrics (ML dataset). (B) Molecular fingerprints—the 7 fingerprints generate a different similarity score for the pairs of drug molecules compared. The median value of each is represented in the box plot (in the center) and the spread shows the density of the drug pairs around that score. (C) MCS—there are two types of scores reported by the MCS algorithm, one is the Tanimoto score and the other is the Overlap coefficient (OC). The violin plots were smoothed for density by an adjustment factor of 3. (D-F)—Performance on the test set. (D) performance of the four models, viz., regularized logistic regression (L1R and L2R), naïve bayes (NB) and random forest (RF) on independent test set for all 5 split-sets. Performance was evaluated using balanced measures: F1 score, matthews correlation coefficient (MCC), positive predictive value (PPV) and area under the curve (AUC). RF clearly had higher performance as compared to the logistic regression and naïve bayes models under all metrics and data splits. The performance of all models was also evaluated using (E) precision-recall and (F) ROC curve–the RF models achieved an AUC of 0.90 averaged on the all 5 test-split sets whereas NB and LRs performed relatively poor on all split-sets (average: NB: 0.68, L1R: 0.51 and L2R: 0.50). (G) High ranking features of RF models on the 5 split-sets–top features are displayed, showing most of the distance-based features provided maximum information gain with ‘Featmorgan’ performing best.