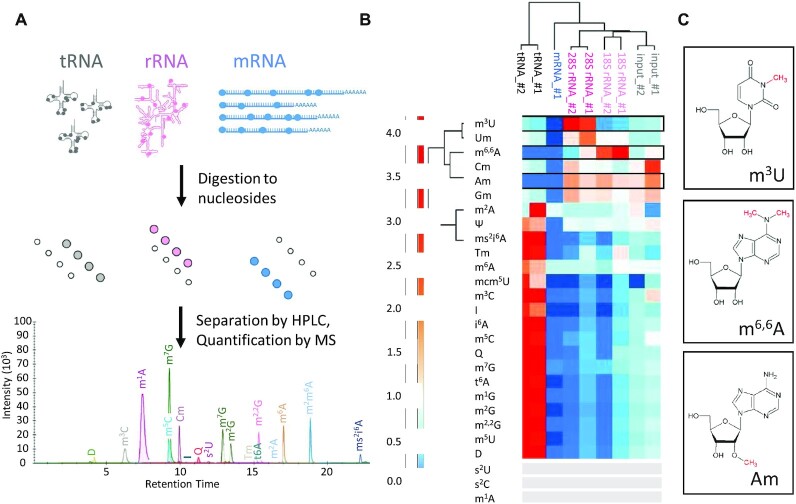
Figure 1.
Distinct modification content of major cellular RNA classes. (A) Workflow of RNA modification measurement. Nucleosides resulting from complete digest of isolated RNA samples were separated by HPLC and quantified by MS/MS. For technical reasons, the presented chromatogram only illustrates 19 out of 27 actually measured modifications (B) Heatmap comparing modification patterns of tRNA (black), mRNA (blue), input (total RNA, grey), 28S rRNA (pink) and 18S rRNA (rose) of mouse liver tissue. n = 3 technical replicates. Two biological replicates (#1) were prepared, except for mRNA; for each RNA type, 3 technical replicates per biological sample were measured. (C) Chemical structure of the candidate modifications for ribosomal markers: m6,6A from 18S rRNA, m3U from 28S rRNA and Am from both rRNA types.