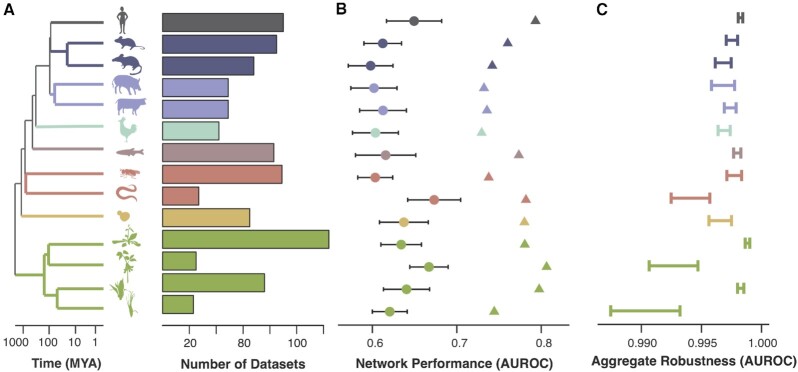
Figure 2.
Aggregate coexpression networks are a powerful tool for comparative genomics. (A) In this work we examine coexpression across species from 3 kingdoms: plants, animals and fungi. Here we focus on the subset of species with GO annotations. The dendrogram shows phylogenetic relationships between these species, and the barplots indicate the number of datasets used to build aggregate coexpression networks. (B) Circles show the mean GO prediction performance for individual networks (+/− standard deviation) while triangles indicate aggregate network performance. (C) Aggregate robustness is high across all species, with variation dependent on n.