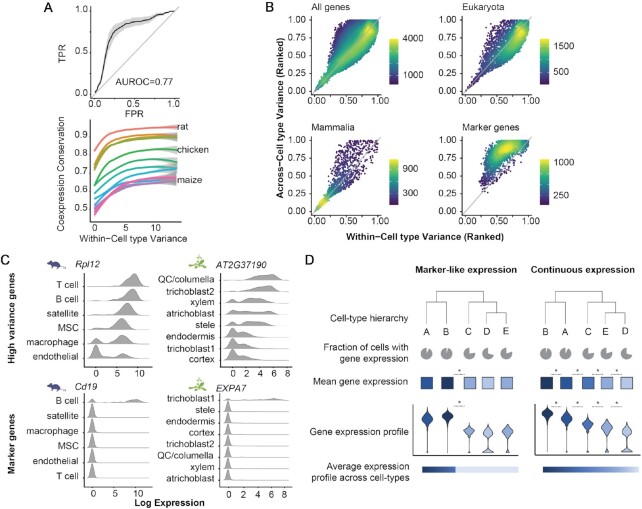
Figure 6.
Genes with conserved coexpression patterns are expressed in continuous gradients across cell types, suggesting an ancient mechanism of cell type divergence. (A) Expression variance is associated with conservation of gene neighborhoods. (Top) Expression variance across > 200 yeast datasets predicts coexpression conservation. TPR = true positive rate, FPR = false positive rate. ROC curves for all species were binned along x-axis, mean +/− SD is plotted. (Bottom) Coexpression conservation is plotted with respect to within-cell type variance in mouse, with loess fits on mean values for each species. (B) Across vs. within-cell type variance in mouse is plotted. Colors indicate local point density. The space can be broken into three regions: genes with high-within and high-across variance are typically more ancient, those that are low/low are more recent, and markers have high-across and low-within cell type variance. Due to their high conservation of coexpression patterns, high/high genes may have conserved functions vis-à-vis cell identity. (C) Examples of continuous (top) vs. marker-like (bottom) expression in mouse (left) and Arabidopsis (right). MSC = mesenchymal stem cell, QC = quiescent center. The conserved gene with high within- and across-cell type variance is more continuous across cell types, whereas the markers (high/low) are either on or off. (D) Schematic illustrating the difference between marker-like and continuous expression across cell types. See Supplementary Figure S7 for additional discussion.