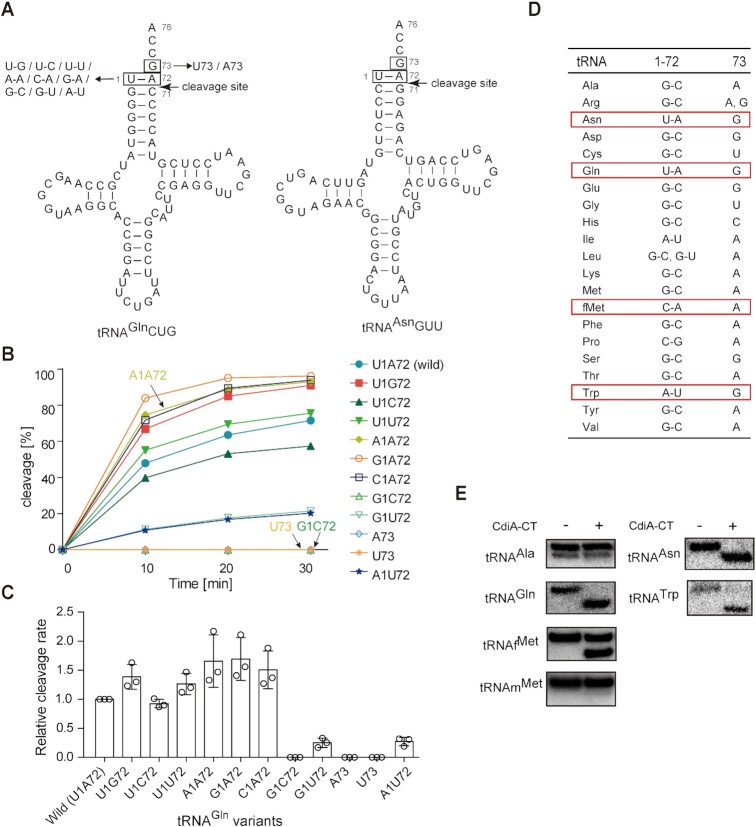
Figure 2.
Recognition elements in tRNA for cleavage by CdiA–CTEC869. (A) Nucleotide sequence and secondary cloverleaf structure of E. coli tRNAGln and tRNAAsn. The mutations introduced into the tRNAGln transcript are depicted on the left. (B) Time course of the cleavage of tRNAGln variants in (A) by CdiA–CTEC869 under neutral pH conditions (pH 7.4). tRNAGln transcript variants (1.0 μM) were incubated at 37°C with 0.2 μM CdiA–CTEC869 in the presence of Tu (0.2 μM), Ts (0.2 μM) and GTP (1 mM). The fraction of cleaved tRNA was quantified at the indicated time points as in (A). (C) Relative cleavage of tRNAGln variants by CdiA–CT in the presence of Tu, Ts, and GTP as in (B), at 37°C. Cleavage level of wild-type tRNAGln at 10 min was taken as 1.0. The error bars in the graphs represent SDs of at least three independent experiments, and the data are presented as mean values ± SD. (D) Comparison of nucleotide compositions at positions 1, 72, and 73 of 20 kinds of tRNAs in E. coli. (E) Cleavage of total tRNAs prepared from E. coli by CdiA–CT in vitro. Total tRNA mixtures (0.5 μg) were incubated at 37°C for 60 min with 0.5 μM CdiA–CTEC869 (20 μl reaction solution) in the presence of Tu (0.5 μM), Ts (0.5 μM) and GTP (1 mM). Cleavage of specific tRNAs was detected by northern blotting using DNA oligonucleotides specific for the corresponding tRNAs (tRNAAla, tRNAAsn, tRNAGln, tRNATrp, tRNAfMet and tRNAmMet).