Figure 7.
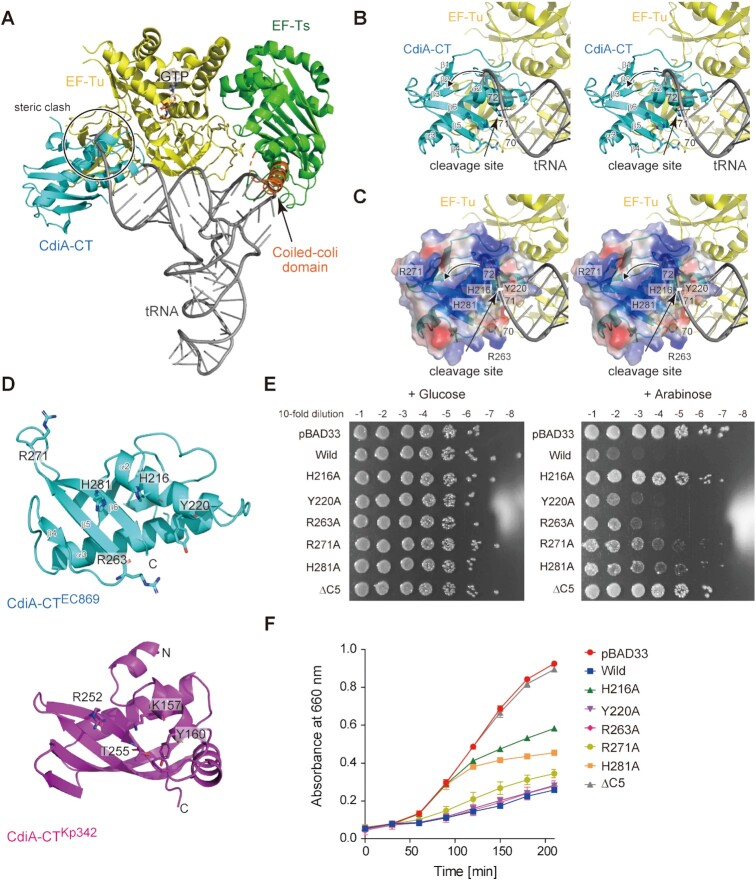
Recognition of the 3′-acceptor region of tRNA by CdiA–CTEC869. (A) A model of tRNA docking onto the CdiA–CT:Tu:GTP:Ts structure. CdiA–CT, Tu, and Ts are colored cyan, yellow, and green, respectively. The coiled-coil domain of Ts is colored orange. tRNA is shown in a stick model (gray). Domains II and III of Tu in the CdiA–CT:Tu complex structure were superimposed onto those in the Tu:GTP:Ts:aa-tRNA complex (Figure 5F), and the CdiA–CT:Tu:GTP:Ts:aa-tRNA model structure was constructed. (B) Close-up view of interactions of 3′-acceptor region of tRNA with CdiA–CT and Tu in (A). The 3′ portion of the tRNA sterically clashes with CdiA–CT. The arrowhead indicates the relocation of 3′-portion of tRNA into the catalytic site for tRNA cleavage. (C) The electrostatic surface potential of CdiA–CT. The positively charged area (blue) resides on the opposite side to the interface between Tu Domain II and CdiA–CT. (D) Comparison of structures of CdiA–CTEC869 and CdiA–CTKp342 from Klebsiella pneumoniae. CdiA–CTEC869 and CdiA–CTKp342 are colored cyan and magenta, respectively. The key amino acid residues required for toxicity and tRNase activity in CdiA–CTKp342 (K157, Y160, R252 and T255) are shown as stick models (16). (E) Effects of mutations on toxicity of CdiA–CTEC869 expressed in E. coli MG1665, as in Figure 3A. LB agar plates containing 50 μg/ml chloramphenicol and supplemented with 0.5% (w/v) arabinose (right panel: +Arabinose) or with 1% (w/v) glucose (left panel: + Glucose). (F) Growth curves of E. coli MG1655 transformed with pBAD33, pBAD33_CdiA–CTEC869 and its variants in LB containing 50 μg/ml chloramphenicol and supplemented with 1% (w/v) arabinose.