Figure 1.
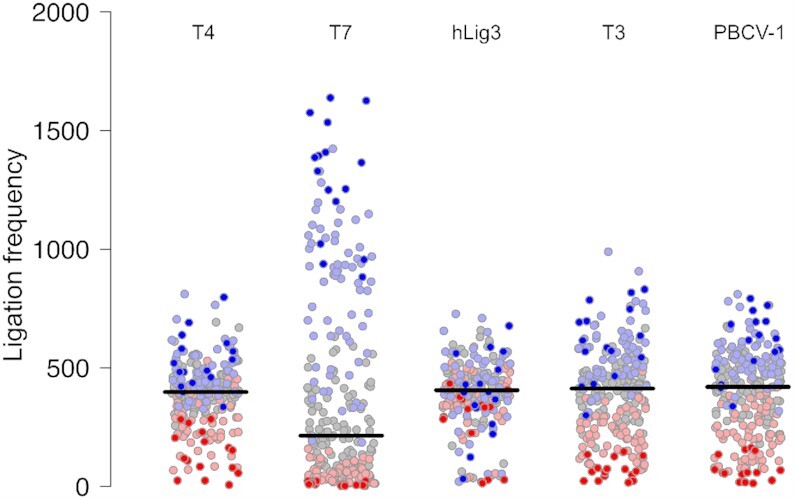
Ligation bias for T4 DNA ligase, T7 DNA ligase, hLig3, T3 DNA ligase and PBCV-1 DNA ligase. The normalized ligation frequency of each overhang was generated by SMRT sequencing of ligation reactions with 100 nM of the multiplexed four-base overhang substrate and 1.75 μM T4 DNA ligase, T7 DNA ligase, hLig3, T3 DNA ligase, or PBCV-1 DNA ligase incubated 1h at 25°C in standard ligation buffer (Supplementary Figures S2A-S2E and File S1 for raw data).T7 DNA ligase exhibits a significant ligation bias with few overhangs ligated very efficiently, and the majority of overhangs ligated with much lower efficiency. T4 DNA ligase and hLig3 exhibit more uniform ligation bias compared to T7 DNA ligase. GC-rich overhangs tend to ligate more efficiently compared to AT-rich overhangs. Each overhang is colored according to its GC content (0% – dark red, 25% – light red, 50% – gray, 75% – light blue, 100% – dark blue.
