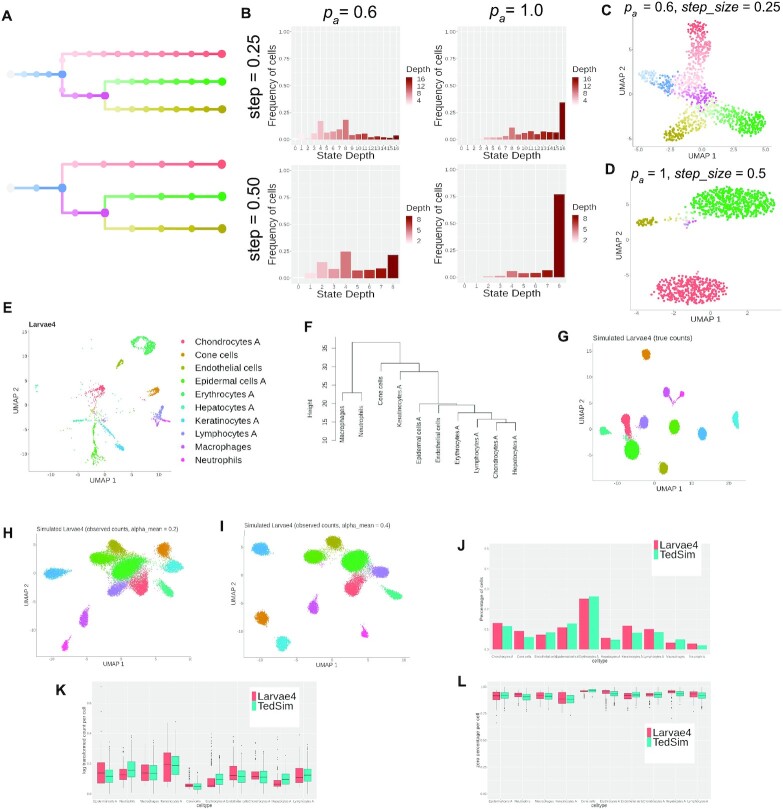
Figure 3.
TedSim simulates both continuous and discrete populations and can generate datasets with multiple cell types that resemble real datasets. (A) Cell state trees sampled with two different step_size =0.25 and 0.5; colours represent cell states in panels (C) and (D). (B) The frequency of cells belonging to each cell state in the simulated dataset, stratified by the depth of the corresponding state in the cell state tree. (C) 2D UMAP visualization of a continuous population of cells where pa and step_size are small, and the trajectory of the states can be observed. (D) 2D UMAP visualization of a discrete population of cells where pa and step_size are large, and the cells are separated into discrete clusters. (E) UMAP visualization of 5-dpf zebrafish larvae cells from (24). Nine thousand three hundred ninety-three cells of 10 cell types are sampled from the original dataset. (F) Cell state tree estimated from the real dataset. Hierarchical clustering is applied to the pairwise distances of the average expressions of the 10 cell types. (G–I) UMAP visualization of simulated zebrafish larvae cells. Twenty simulations each with 1024 cells are generated with selected parameters to fit the dataset to the referred real dataset. (G) True counts without technical noise. (H and I) Observed counts with different capture efficiency values. (J) Comparison of cell type compositions between simulated data and real data. (K and L) Statistical comparison between cell types of simulated data and the real data. Both metrics are calculated based on observed counts with capture efficiency alpha_mean = 0.1. Each box corresponds to a cell type. Red boxes represent real data and green boxes represent TedSim simulated data. (K) Average expression per cell. (L) Average zero percentages per cell.